| Date of Received | 02 July 2019 |
| Date of Revision | 13 February 2020 |
| Date of Acceptance | 21 February 2020 |
| Indian J Pharm Sci 2020;82(2):300-305 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of the present study is to evaluate the role of phytoconstituents of Ganoderma lucidum in the interactions with non-metastatic clone 23, isoform H1. 3D structure of non-metastatic clone 23, isoform H1 was retrieved from the PDB data bank with further optimization of both the protein and ligands. In silico binding potential of the selected ligands with non-metastatic clone 23, isoform H1 was determined using the Auto Dock Tool 2.0 and visualized with Bio via discovery studio visualizing tool to assess the molecular binding properties of the ligands with non-metastatic clone 23, isoform H1 by molinspiration calculations and further assessment of drug likeliness. Ganoderic acid H appeared to possess promising binding with non-metastatic clone 23, isoform H1 with least binding energy of –8.87 Kcal/mol forming 5 hydrogen bond interactions followed by ganoderic acid T and A. The findings of this study thus provide theoretical evidence for ganoderic acid derivatives from Ganoderma lucidum to possess promising binding to non-metastatic clone 23, isoform H1 suggesting it as a target to arrest tumour metastasis with further in vivo validation
Keywords
NM23-H1, Ganoderma lucidum, tumour metastasis, hepatitis
Hepatocellular carcinoma (HCC) has increased substantially with an annual incidence rate of 9% in the last two decades globally, which is attributed to hepatitis C virus (HCV) widely transmitted through blood to blood contact[1]. Major global health burden persists due to the progression of acute hepatitis to liver cirrhosis and carcinoma, with poor prognosis due to the increased risk of metastasis. Various host encoded markers and proteins are associated with the progression of HCC and its metastasis. Amidst various host encoded proteins, non-metastatic clone 23, isoform H1 (NM23-H1) is a metastasis suppressor gene against HCV and has been in association with a wide variety of oncogenic tumours in humans[2]. NM23-H1 gene encodes for a protein that catalyses the phosphorylation of nucleoside diphosphates to nucleoside triphosphates and its reduced expression can enhance metastasis of tumours in host tissue[3]. NM23-H1 expression levels are highly altered based on the host-virus interactions and is considered to be modulated by various tumour associated genes in enhancing metastasis with further tumour or cancer progression in HCC and in other viral-associated cancers[4]. In addition, NM23-H1 in association with other co-located cellular proteins also do induce variations in cellular migration, cell adhesion, apoptotic and necrotic functions etc.[5]. As a contradictory overexpression of NM23-H1 also induces a high progression of cervical cancer in correlation with other associated parameters[6]. It has been thus documented NM23-H1 could be a novel candidate to target the regulation of cancer metastasis, and especially in HCV mediated HCC. Novel antiviral drugs are found to be clinically ineffective to specifically target NM23-H1, thus exploring the need of alternative compounds as ligands in their regulations.
In this view, the present study is aimed to target NM23-H1 with the constituents of the mushroom,
G. lucidum, which has been documented to be a traditional tropical country medicine with various medicinal properties. G. lucidum is a white rot fungus and is popularly used in Chinese medicine, contains many triterpenoids in the form of ganoderic acids and reported to be cytotoxic to human carcinoma cell lines but with less toxicity to normal cell lines in a dosedependent manner[7]. G. lucidum also enhances natural killer cell activity by inducing high level secretion of perforins and granulysins[8]. The protective effects of G. lucidum extract is also reported against a wide variety of liver injuries[9]. This investigation is aimed at assessing the NM23-H1 enhancing role of ganoderic acids T, A and H from G. lucidum using in silico docking analysis.
Materials and Methods
Retrieval of NM23-H1 and protein optimisation:
The crystal structure of the metastasis inhibition factor NM23-H1, the tumor metastatic inhibition process associated protein was retrieved from RCSB (Research Collaboration for Structural Bioinformatics) protein data bank (PDB, http://www.rcsb.org/pdb). The protein was prepared by removing the co crystal ligands, water and co-factors from the downloaded crystal structure. Hydrogen atoms, solvation parameters and fragmental volumes to the protein were added and electronic charges were assigned to the protein atoms using Kollman united atoms force field by using Auto Dock Tool (ADT)-2.0.
Ligand preparation and optimisation:
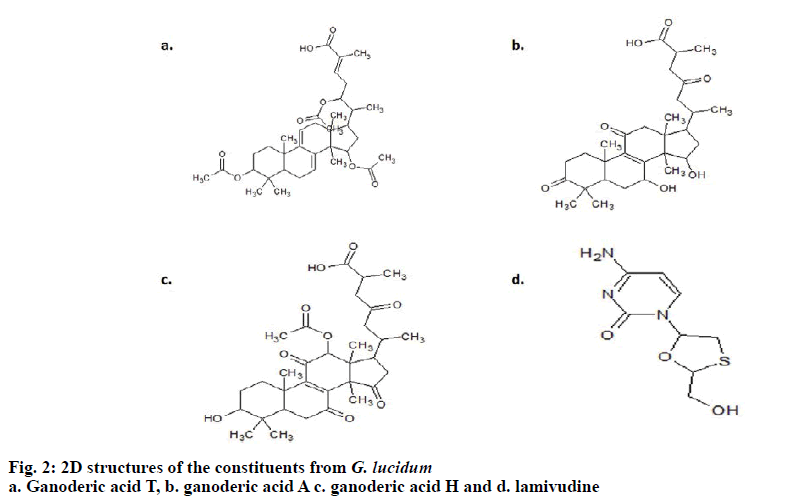
Using Chemsketch Software, the structures of ganoderic acid T, ganoderic acid A, ganoderic acid H and lamivudine were drawn together with their 3-D structures and optimization. The selected ligands were retrieved in SDB format, which were further saved in .mol file followed by the subsequent conversion using open babel molecular converter program[10] and were saved in PDB format.
Docking simulations and interpretations:
The docking analysis to interpret the affinity between ganoderic acids T, A, H and lamivudine with NM23-H1 in oncogenic hepatitis viruses was achieved by ADT with the intermediary steps such as pdb.qt files for proteins and ligands. Using graphical user interface program ADT, the grid box creation was completed. Prior preparation of the grid map using the grid box with a grid size of 126×126×126 xyz points was done.
Further using Lamarckian genetic algorithm (LGA), docking simulation was achieved by setting the initial position, orientation and torsions of the ligand molecules in a random position. Ten different runs set to terminate after a maximum of 250 000 energy evaluations was used for each docking experiment with the population size set at 150. A translational step of 0.2 Å, quaternion and torsion steps of 5 were applied for each dock. The most favourable free energy of binding is achieved by clustering the results >1.0 Å in positional root-meansquare deviation (RMSD)[11]. Finally, the pose was extracted and aligned with the receptor structure with the lowest binding energy or binding affinity for final analysis.
Docking visualisation:
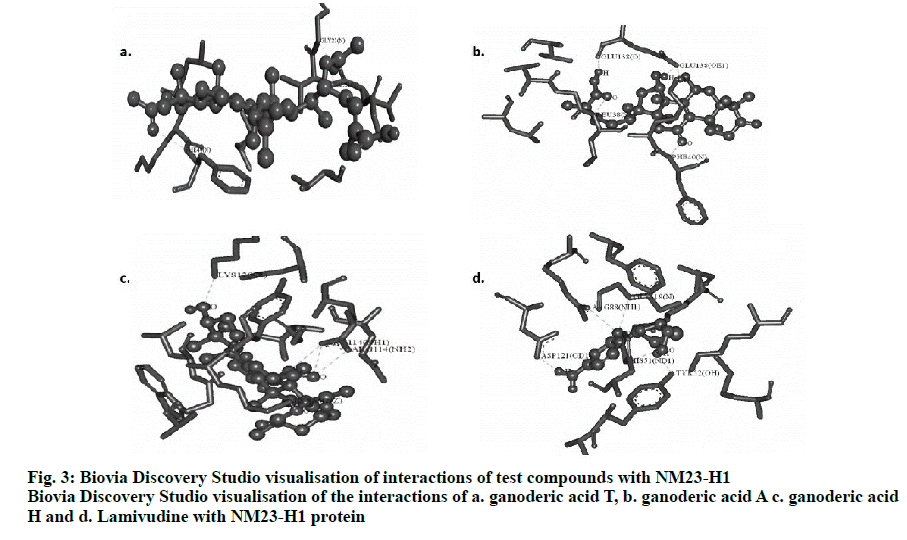
The protein-ligand interactions like hydrogen bonding and other non-bonded energies between the ganoderic acids T, A, H and the lamivudine with NM23-H1 were visualized using Biovia Discovery Studio Visualizer software. The relative stabilities were evaluated using their molecular dynamics, binding affinities, energy simulations with further docking score assessments.
Results and Discussion
The 3D crystal structure of human nucleoside diphosphate kinase A (NM23-H1) was downloaded from PDB database and its PDB structure id was documented as 1UCN–A chain (strain SC5314). Removal of the water molecules and final stage merging of hydrogen atoms to the receptor molecule was successful. The 3D structure was visualized using RasMol with the analysis of pink colour indicating the alpha-helix, yellow arrow indicating the beta sheets and white colour indicating the turns (fig. 1). Ligand optimization was achieved using ACD Chemsketch and retrieved in a compatible format using Open Babel molecular converter tool. 2D and 3D structures of the ligands and its SMILES format were obtained successfully (fig. 2).
The best conformers were selected using LGA based on the best ligand-receptor structure from the docked structure based on the lowest energy and minimal solvent accessibility. Biovia discovery studio visualizing tool of the hydrogen bond interactions in 3D and 2D stick model between the ganoderic acids T, A, H and lamivudine with NM23-H1 is given in fig. 3. The amino acids of NM23-H1 binding with the bioactive compounds, showed ganoderic acid H with the lowest estimated free binding energy of –8.87 Kcal/mol with 5 hydrogen bond interactions, followed by ganoderic acid T with a binding energy of –6.78 Kcal/mol with 4 hydrogen bond interactions and ganoderic acid A with a binding energy of –6.06 Kcal/mol with 5 hydrogen bond interactions in comparison to lamivudine. The torsional energy and the docking scores between the drug and ligands are also documented in Table 1. The inter-molecular interactions are recorded in Table 2. The docking results shows Ganoderma sp., constituents with good binding energy with the target receptor with NM23-H1 in comparison with lamivudine.
| NM23-H1 docking with | Hydrogen bonds | Binding energy | Ligand efficiency | Intermolecular energy | vdW+Hbond+desolv Energy | Electrostatic energy | Torsional energy | Total internal Unbound |
|---|---|---|---|---|---|---|---|---|
| Ganoderic acid T | 4 | –6.78 | –0.15 | –10.78 | –10.36 | –0.11 | 3.58 | –2.29 |
| Ganoderic acid A | 5 | –6.06 | –0.16 | –8.75 | –8.62 | –0.12 | 2.68 | –2.18 |
| Ganoderic acid H | 5 | –8.87 | –0.22 | –11.85 | –10.16 | –1.69 | 2.98 | –2.65 |
| Lamivudine | 5 | –4.47 | –0.3 | –5.66 | –5.48 | –0.18 | 1.19 | –1.14 |
Table 1: Docking Results of Ganoderic Acids T, A, H and Lamivudine with Nm23-H1
| NM23-H1 docking with | Hydrogen bond interactions | van der Waals interactions | π-σ interactions | alkyl/π-alkyl interactions | π-sulfur interactions |
|---|---|---|---|---|---|
| Ganoderic acid T | 4 (GLY22, VAL21, PHE40, LYS39) | 2 (LEU20, GLY19) | 1 (PHE40) | 13 (VAL16, VAL21, VAL74, PHE40, LEU38) | - |
| Ganoderic acid A | 5 (GLU138, PHE40, LEU38) | 3 (GLY37, VAL36, VAL140) | - | 4 (LYS39, LEU35, LEU38) | - |
| Ganoderic acid H | 5 (LYS12, ARG114, LYS66) | 4 (GLY63, GLY113, GLN111, ASN115) | - | 6 (TRP60, VAL112, TYR67, LEU64) | - |
| Lamivudine | 5 (ARG88, GLY119, TYR52, HIS51, ASP121) | 6 (ARG105, LEU55, GLY118, LYS12, PHE8, MET68) | - | 1 (TYR52) |
Table 2:Interactions of Ganoderic Acids T, A, H and Lamivudine with Nm23-H1
Expression of NM23-H1 and its functions are known to modulate the tumour-associated metastasis and further progression especially in viruses like Kaposi’s sarcoma associated herpes virus (KSHV), Epstein Barr virus and human papilloma virus (HPV) mediated cancers[12].
NM23-H1 gene is considered as a potent factor as it encodes protein products for tumour suppression and metastasis in different forms of tumours especially in oncogenic hepatitis viruses. NM23-H1 plays a vital role in varying the extent of tumour progression and further metastasis[13]. Until now, as there are no clinically effective antiviral drugs to inhibit tumour metastasis, NM23-H1 is considered as the best target for activation by natural compounds. In this context, G. lucidum, a mushroom containing ganoderic acids was known to exert toxic effects against viruses[14]. Possible mechanism of G. lucidum antiviral effects on herpes group of viruses is already documented[15]. In addition, G. lucidum is also reported to possess a promising role in treating viral hepatitis[16]. However, the best fit of the G. lucidum constituents with the NM23-H1 and to enhance its tumour suppression and metastasis in various forms of cancers especially in HCV-associated tumours has not been studied. In the present investigation, this activity was assessed effectively using molecular docking analysis, which is a molecular modelling technique. Amidst many tumour suppressor proteins, NM23-H1 was retrieved from the PDB database as a desirable target based on the data recorded in database and was freely accessible as nucleoside diphosphate kinase A 1UCA chain. In the present study, ganoderic acids T, A, H and lamivudine was docked with NM23-H1 that resulted in a promising receptor-ligand complex. Docking analysis involved 2 major steps where the prediction of the exact orientation of the conformers in to the best active site pocket called pose and the strength of target-ligand binding interactions is achieved by scoring[17]. The best docked ligand models were selected according to the lowest binding energy that comprised the largest conformational cluster in each binding. ADT-4.2, which is considered as a suite of automated docking tool with a software for modelling flexible small molecule such as drug molecule binding to receptor proteins of known 3D structure was used. Using genetic algorithms for the conformational search, a rapid grid-based method of energy evaluation was achieved in the study to analyse the docking simulations. The relative binding affinities for ganoderic acid derivatives and lamivudine helped to observe the structure-inhibitory action relationships. In this study, the LGA was used to explore the binding conformational landscape of ganoderic acids T, A, H and lamivudine with NM23-H1. The docking scores on NM23-H1 indicated that there was a direct relationship between the energy of the binding affinity, referring to the lowest docking scores and the stability. In accordance with this, apart from the binding energy, the inter-molecular energy, van der Waal’s energy and torsional energy were also at a higher end for ganoderic acid H but with highest alkyl/π-alkyl interactions for ganoderic acid T.
Two and three dimensional conformational structures of the ligand-protein complexes were visualized using the Biovia Discovery Studio Visualizer to investigate the binding modes. Analysis predicted the hydrogen bond interactions between NM23-H1 and the ligands, yielding promising results with hydrogen bonds and bonding energies. The number of hydrogen bonds together with the enthalpic gain due to the water molecules determines the best fit[18]. In this context, by assessing the binding energy and their corresponding ligand efficiency, intermolecular energy, electrostatic energy, vdW+Hbond+desolv energy, internal energy and torsional energy of NM23-H1 with the ganoderic acid derivatives and lamivudine, showed ganoderic acid H to be the best enhancing agent of NM23-H1 suggesting its role to inhibit or arrest the tumour metastasis, which could be useful in controlling hepatitis-associated carcinomas. This is followed by ganoderic acid T and ganoderic acid A in comparison to lamivudine, which yielded less energy of -4.47 Kcal/mol but with 5 hydrogen bonds as that of ganoderic acid H.
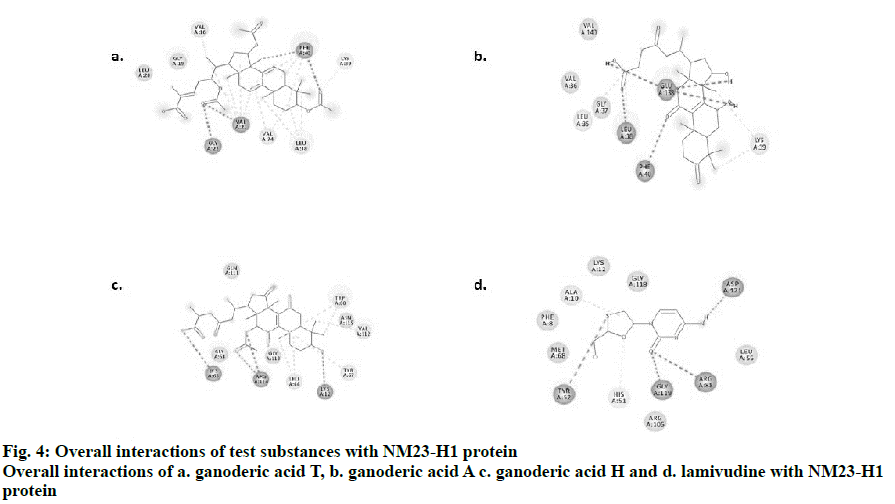
Visualization by Biovia Discovery studio tool enabled us to view the 2D structures of the drug-ligand interactions. The ligands, ganoderic acids T, A, H and lamivudine bind to NM23-H1 through non-covalent interactions such as H-bonding, alkyl, alkyl-π, π-π, π-σ and van der Waals interactions. Simplified visualization was achieved and is illustrated in 2D, with H-bonding as dark green circles associated with green dashed lines, van der Waals forces as medium light green circles, carbon-oxygen dipole-dipole interaction as light green circles with dashed lines, alkyl-π interactions as light pink circles with dashed lines, T-shaped π-π stacking and parallel π-σ stacking as both indicated with dark pink circles (fig. 4). The present investigation provided theoretical evidence for designing novel drugs from the mushroom G. lucidum using computational bioinformatics tools and databases[19,20], which could be effective against tumour metastasis especially in viral hepatitis by targeting NM23-H1. Selection of tumour suppression enhancing compounds from G. lucidum was achieved in the present study using in silico molecular docking assessments. The docking evaluation in this study suggested promising role for 3 different forms of ganoderic acids T, A and H in enhancing tumour suppression and further metastasis with promising binding to NM23-H1. Other associated interactions like interactions with ADP might also be possible with these compounds, which requires further confirmation. Experimental validation in vivo and in vitro would further substantiate the theoretical evidence obtained from this study. This preliminary evidence would promote further target-based experimental screening of the G. lucidum constituents for a highly selective mechanism of action to prevent metastasis in viral hepatitis.
References
- Altekruse SF, Mcglynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality and survival trends in the United States from 1975 to 2005. J Clin Oncol 2006;27(9):1485-91.
- Steeg PG, Bevilacqua G, Pozzati R, Liotta LA, Sobel ME. Altered expression of NM23 a gene associated with low tumor metastatic potential during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Res 1988;48(22):6550-4.
- Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomemb 2000;32(3):301-8.
- Kaul R, Murakami M, Kumar P, Robertson ES. Nm23 as a Metastasis inhibitor In: Thomas-Tikhonenko A, editor. Cancer Genome and Tumor Microenvironment. New York, NY, USA: Springer 2010;233-71.
- She S, Xu B, He M, Lan X, Wang Q. Nm23-H1 suppresses hepatocarcinoma cell adhesion and migration on fibronectin by modulating glycosylation of integrin beta. J Exp Clin Cancer Res 2010;29:93-7.
- Munoz N, Bosch FX, De Sanjose S, Herrero R, Castellsague X, et al. International agency for research on cancer multicenter cervical cancer study. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003;348(6):518-27.
- Ruan W, Wei Y, Popovich DG. Distinct responses of cytotoxic Ganoderma lucidum triterpenoids in human carcinoma cells. Phytother Res 2015;29(11):1744-52.
- Chang CJ, Chen YY, Lu CC, Lin CS, Martel J, Tsai SH, et al. Ganoderma lucidum stimulates NK cell cytotoxicity by inducing NKG2D/NCR activation and secretion of perforin and granulysin. Innate Immun 2014;20(3):301-11.
- Yihuai Gao, He Gao, Eli Chan, Wenbo Tang, Xiaotian Li, Jun Liang. Protective effect of ganoderma a mushroom with medicinal properties against various liver injuries. Food Rev Int 2007;6:27-52.
- Boyle NM, Banck M, James CA, Morleyc, Vandermeersch T. Open babel an open chemical toolbox. J Cheminform 2011;3:33.
- Blum C, Blesa MJ, Roli A, Sampels M. Hybrid meta heuristics an emerging approach to optimization. Studies in Computational Intelligence. Heidelberg: Springer-Verlag 2008;114.
- Rickinson AB, Kieff E. Epstein-Barr virus In: Fields BN, Knipe DM, Howley PM. editors. Fields Virology, 3rd ed. Philadelphia, PA,USA: Lippincott-Raven Publishers 1996;65.
- Hwang BG, Park IC, Park MJ, Moon NM, Choi DW, Hong WS, et al. Role of the NM23-H1 gene in the metastasis of gastric cancer. J Korean Med Sci 1997;6(12):514-8.
- Weinjing Z, Xiao PY, Zhuliang Y, Lizhang, Hon SG, Kai LW, et al. Antiviral effects of two Ganoderma lucidumtriterpenoids against enterovirus 71 infection. Biochemical and Biophysical Research Communications 2014;449(3):307-12.
- Seong K, Young S, Kim C, Kilee S, Sun H. Possible mode of antiviral activity of acidic protein bound polysaccharide isolated from Ganoderma lucidum on herpes simplex viruses. J Ethnopharmacol 2000;72(3):475-81.
- Yihuai G, Shufeng Z, Min H, Anlong X. Antibacterial and antiviral value of the genus Ganoderma P. Karst. Species a phyllophoro mycetideaea: A Review. Int J of Med Mushrooms 2010;5(3):235-46.
- Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 2003;46:2287-303.
- Clarke C, Woods RJ, Gluska J, Cooper A, Nutkey MA, Boons GJ.Involvement of Water in Carbohydrate-Protein Binding: Concanavalin A Revisite. J Am Chem Soc 2001;123:1-6.
- Vijayashree PJ, Smiline GAS, Paramasivam A. Enterococcus faecalis: an emerging microbial menace in dentistry-an insight into the in-silico detection of drug resistant genes and its protein diversity. J Clin Diag Res 2018;12(10):106.
- Vijayashree PJ, Smiline GAS, Paramasivam A. An in-sight into the emergence of Acinetobacter baumannii as an oro-dental pathogen and its drug resistance gene profile: An in-silico approach. Heliyon 2018;4(12):e01051.