- *Corresponding Author:
- X. M. Du
Department of Maxillofacial Surgery, Tianjin Stomatological Hospital, School of Medicine, Nankai University, Tianjin 300041, China
E-mail:15620802391@163.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “144-151” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Oral squamous cell carcinoma is the most common malignancy of the oral neoplasm, comprising for more than 90 % of all oral malignancies, as well as approximately 38 % of head and neck tumors. So, it is of great significance to find the potential therapeutic target of the disorder. The messenger ribonucleic acid and micro ribonucleic acid expression profiling was performed for oral squamous cell carcinoma patients; micro ribonucleic acid-messenger ribonucleic acid regulatory network was established; the functions of potential micro ribonucleic acid and messenger ribonucleic acid were further explored in Cal27 oral adenosquamous carcinoma cell line. Compared with normal mucosa, oral squamous cell carcinoma patients demonstrated 192 differentially expressed genes (66 were up-regulated and 126 were downregulated) and 23 differentially expressed micro ribonucleic acids (4 were up-regulated and 19 were downregulated), among which micro ribonucleic acid-196a and annexin-A1 were significant and differentially expressed. The annexin-A1 was a direct target of micro ribonucleic acid-196a supported by bioinformatics and luciferase analysis. The antagomiR-196a technique was utilized to silence the expression activity of micro ribonucleic acid-196a. By this, micro ribonucleic acid-196a was shown to modulate oral squamous cell carcinoma cellular migration, invasion and colony formation through annexin-A1. At the same time, the functions of micro ribonucleic acid-196a and annexin-A1 were closely associated with protein kinase B and mitogen-activated protein kinase signaling pathways. This integrated study hypothesized a micro ribonucleic acid-196a dependent signaling axis for oral squamous cell carcinoma and provided a beneficial reference for future clinical study.
Keywords
Oral squamous cell carcinoma, micro ribonucleic acid-196a, annexin-A1, mitogen-activated protein kinase
Head and Neck Squamous Cell Carcinoma (HNSCC) represents a heterogeneous collection of malignancies for the upper aerodigestive tract, salivary glands and thyroid[1], which has emerged as the 6th leading cancer by incidence and 8th by death worldwide[2,3]. The Oral Squamous Cell Carcinoma (OSCC) is the primary malignancy of HNSCC, accounting for nearly 95 % of all HNSCC patients[4]. As estimated, the annual incidence of OSCC is over 300 000 cases[5]. Meanwhile, the worldwide incidence of OSCC is not homogeneous based on the variable prevalence of the risk factors, with a relative higher prevalence in South East Asia, Brazil as well as central Europe and 62 % of annual incidence arising in developing countries[6]. Growing evidences from multiple retrospective assessments have suggested the connections between OSCC and environmental risk factors including diabetes, alcoholism, smoking as well as dietary habits[7,8]. However, even with decades of study, the knowledge of the genetic basis for OSCC is far from satisfaction, which is imperative for early detection, treatment, as well as the improvement of patient survival. In 2011, researcher demonstrated the top 10 primary alterations and fundamentals for OSCC formation and development, which are sustaining proliferative signaling, evading growth suppressors, avoiding immune destruction, activating invasion and metastasis, tumor-promoting inflammation, enabling replicative immortality, inducing angiogenesis, genome instability and mutation, resisting cell death, as well as deregulating energetics[9].
The micro Ribonucleic Acids (miRNAs) are small non-coding RNAs (ncRNAs) with 21-25 nucleotides in length, which are highly conserved among distinct species[10]. Since it was discovered in 1993, miRNA was found in all eukaryotic cells conserved across the species, which is functional via gene silencing and translational repression by binding to target messenger RNAs (mRNAs)[11]. Currently, the miRNAs have been shown to be closely associated with various progressions of distinct human disorders such as cardiovascular disease, neurodegenerative diseases, retinal disorder and several types of cancer[12-14]. For instance, increasing evidence claimed that miR-223 might play an essential part in both hematological malignancies and solid tumors[15]. Meanwhile, miR-34 has been shown to be involved in the regulation of cell cycle and apoptosis through multiple signaling pathways such as tumor protein p53, Retinoic Acid (RA) and Wingless-related integration site (Wnt) signaling in colon cancer[16]. Thus, studies on miRNAs provide a great opportunity to improve our understanding of complex biological mechanisms in carcinogenesis. At the same time, accumulating evidences have suggested that miRNA-based therapies, either restoring or repressing miRNA expression and activity, hold great promise for future cancer study[17]. However, the connections between miRNA-based molecular mechanism and OSCC are uncertain yet.
In the view of pivotal values of miRNAs in cancer prognosis and treatment prediction, we comprehensively explored the potential hub genes (mRNAs and miRNAs) for OSCC patients based on an innovative combination of bioinformatics methods and clinical experimental verification here. The most significant Differentially Expressed miRNA (DEM, miR-196a) as well as the associated signaling cassette were further examined in-depth in this study, which was beneficial for future OSCC therapy.
Materials and Methods
Clinical specimens:
The tissues of participants used in this study were kindly provided by our colleagues. The individuals were confirmed with a histopathological diagnosis of OSCC without any previous history of treatment. The patients were categorized into Grade I, Grade II, Grade III and Grade IV using the traditional World Health Organization (WHO) classification. This study was approved by the institutional review committee.
Tissue sample collection and RNA isolation:
The incision biopsy of tumor tissues was taken from each individual either during the investigative biopsy procedure or surgical excision of the lesion. The normal mucosa was collected from patients undergoing oral and maxillofacial surgery for reasons other than OSCC. All the collected tissue bits were immediately snap frozen and stored in liquid nitrogen. The total RNA was extracted using RNAiso plus (Takara, Beijing, China). The high-throughput sequencing was conducted and analyzed by Genewiz Co., Tianjin based on Applied Biosystems (ABI) solid sequencer platform.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) analysis of mRNA and miRNA expression:
The high-capacity RNA to complementary Deoxyribonucleic Acid (cDNA) kit (Applied Biosystems) was utilized to reversely transcribe RNA (1000 ng) to cDNA and qPCR amplification was performed with SYBR Green kit (Qiagen). The measurements were normalized according to the internal reference gene Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) of each parallel sample. For miRNA expression analysis, the TaqManTM Advanced microRNA assay kit (Applied Biosystems) and miRNA-specific primers reverse transcription RNA (100 ng) were used. All the experiments were performed in triplicate independently.
Prediction of miRNA target genes:
The target genes of miRNAs were predicted by the microRNA Target Prediction Database (miRDB) (http://mirdb.org/index.html, version 6.0)[18]. At the same time, the Cytoscape (https://cytoscape.org/, version 3.7.2) was performed to visualize the miRNA-mRNA regulatory network.
Cell culture:
The Oral Adenosquamous Carcinoma cell line (Cal27) cells were purchased from Chinese Academy of Sciences (Shanghai, China) and cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Hyclone, United States of America (USA)) supplemented with 10 % Fetal Bovine Serum (FBS, Gibco, USA). As for transfection assay, the cells were seeded into a 6-well plate one day before and the LipofectamineTM 3000 (Life Technologies, USA) were used. The mimic of miR-196a (antagomiR-196a) and its corresponding Negative Controls (antagomiR-NC) were designed and synthesized by Life Technologies Corporation. The Annexin-A1 (ANXA1) small interfering RNA (siRNA) and siRNA Negative Control (siRNA-NC) were constructed by Genewiz Corporation (Tianjin, China). The 8 nM MK-2206 dihydrochloride (MK-2206) and 6.1 nM ravoxertinib (MedChemExpress (MCE), USA) were used for signaling pathway analysis.
The luciferase reporter assay:
The 3'-non-coding region of ANXA1 was synthesized and inserted into the XhoI and NotⅠ sites of the pCheck2 reporter luciferase vector downstream of the luciferase gene after annealing. The wild-type or mutant plasmid and negative control or miR-30a was co-transfected into Cal27 cells. The luciferase analysis was performed by dual luciferase reporter analysis system (Promega, USA).
Cell migration and cell invasion assay:
The assays were performed according to the previously established protocol[19]. The scratch assay was used to examine cell migration. Meanwhile, the cell invasion assays were performed with transwell chamber (Costar, USA).
Functional enrichment analysis:
For the obtained Differentially Expressed Genes (DEGs), we used the “clusterProfiler” function package in R language for enrichment analysis of Gene Ontology (GO) (including Biological Process, Molecular Function and Cellular Component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. When p-value<0.05, we considered the corresponding entries to be significantly enriched.
Statistical analysis:
All the experiments were conducted in triplicate independently. The Statistical Analysis System (SAS) 9.4 software was developed for data analysis. The continuous variables were tested for normal distribution and the student t-test in SAS 9.4 was used to analyze the difference between two groups, with p<0.05 considered as a significant difference.
Results and Discussion
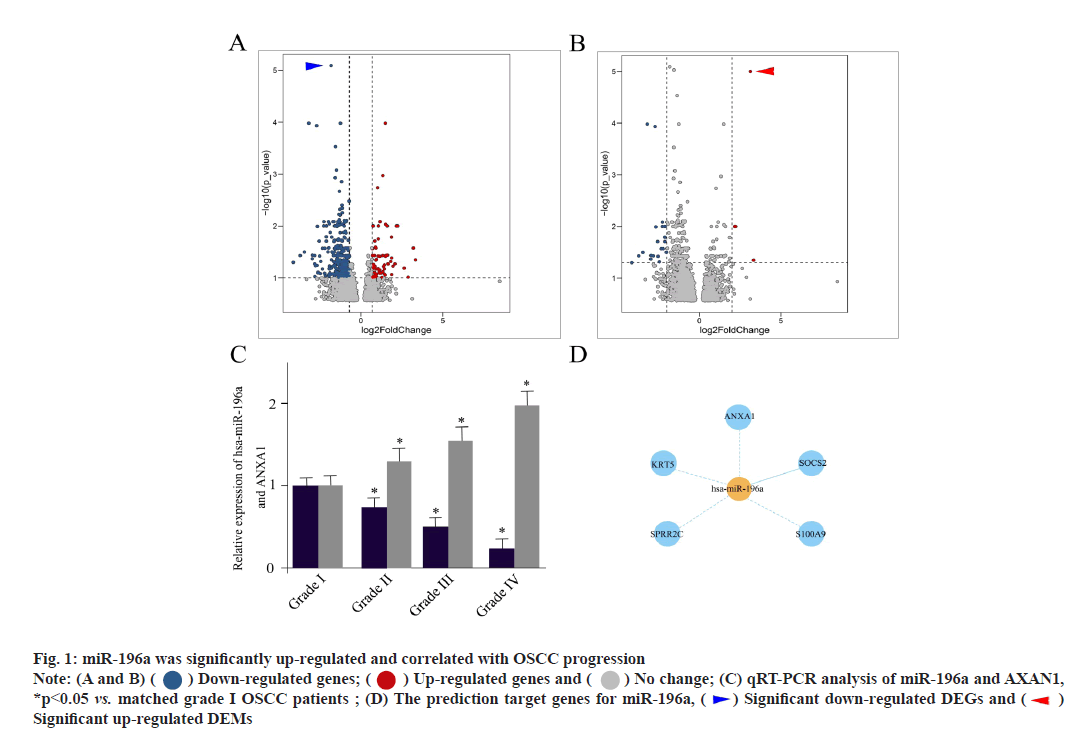
The miR-196a was significantly up-regulated and correlated with OSCC progression. First of all, we aimed to identify DEMs and DEGs (mRNAs) for OSCC patients in respect to normal mucosa. The volcano map of DEGs (mRNAs) between two groups of OSCC patients was shown here. The horizontal axis represents the multiple of differential expression (Log2FC), the vertical axis represents -log10 (False Discovery Rate (FDR)). The blue dots represent down-regulated genes and the red dots represent up-regulated genes respectively. The most significantly down-regulated DEGs were indicated as blue arrowhead (fig. 1A). The volcano map of DEMs between two groups of OSCC patients was described here. The most significantly up-regulated DEMs were indicated as red arrowhead (fig. 1B). In this set of 40 samples including 20 tumors and 20 normal controls, OSCC patients demonstrated 192 DEGs (66 were up-regulated and 126 were downregulated) and 23 DEMs (4 were up-regulated and 19 were down-regulated), as shown in fig. 1A and fig. 1B. The expression level of ANXA1 was significantly decreased in OSCC patient tissues, indicated as blue arrow head in fig. 1A. At the same time, the activity of miR-196a was significantly accelerated, indicated as red arrow head in fig. 1B.
Fig. 1: miR-196a was significantly up-regulated and correlated with OSCC progression
Note: (A and B) ( ) Down-regulated genes; (
) Down-regulated genes; ( ) Up-regulated genes and (
) Up-regulated genes and ( ) No change; (C) qRT-PCR analysis of miR-196a and AXAN1,
*p<0.05 vs. matched grade I OSCC patients ; (D) The prediction target genes for miR-196a, (
) No change; (C) qRT-PCR analysis of miR-196a and AXAN1,
*p<0.05 vs. matched grade I OSCC patients ; (D) The prediction target genes for miR-196a, ( ) Significant down-regulated DEGs and (
) Significant down-regulated DEGs and ( )
Significant up-regulated DEMs
)
Significant up-regulated DEMs
To further assess the clinical significance of miR-196a and ANXA1, we grouped all OSCC samples based on the traditional WHO classification. The qRT-PCR analysis of miR-196a and AXAN1 between different grades of OSCC patients was shown here. It could be shown that the increased miR-196a expression was positively associated with OSCC progression. While, the expression pattern of ANXA1 was opposite (fig. 1C). The prediction target genes for miR-196a were shown in fig. 1D. The target of miR- 196a was established using miRDB, TargetScan and miRTarBase, which suggested ANXA1, Keratin 5 (KRT5), Small Proline-Rich Protein 2C (SPRR2C), Suppressor of Cytokine Signaling 2 (SOCS2) and S100 Calcium Binding Protein A9 (S100A9) were the potential targets (fig. 1D). These outcomes implied that miR-196a participated in the progression of OSCC.
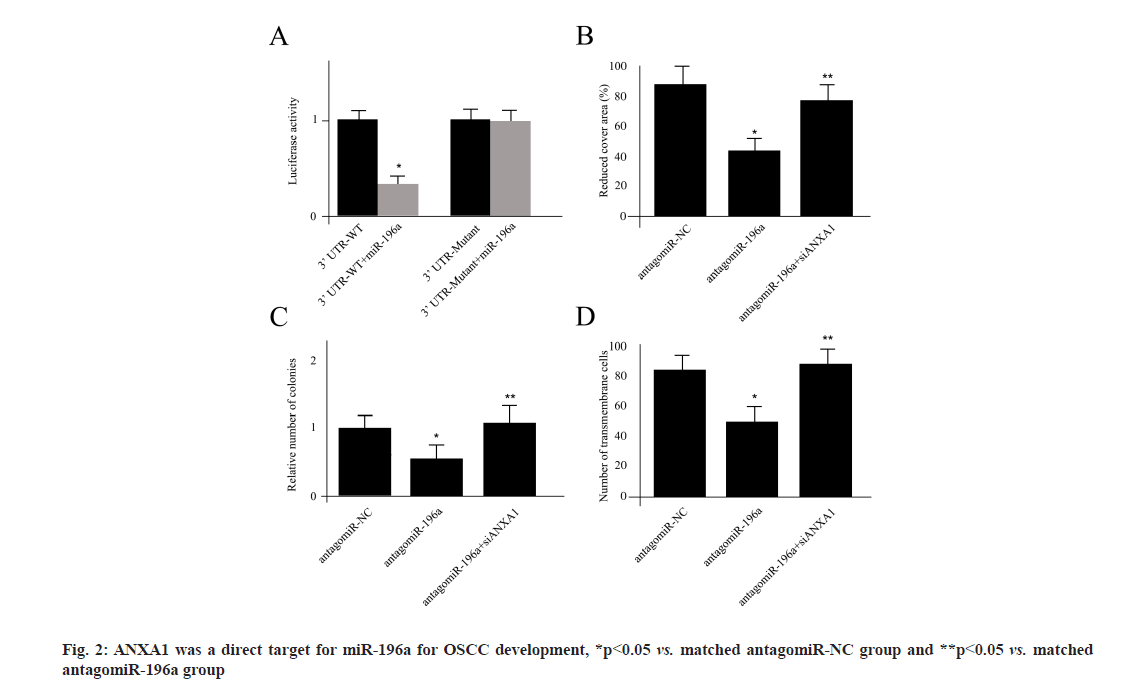
ANXA1 was a direct target for miR-196a for OSCC development. Next, we sought to verify whether ANXA1 was a direct target of miR-196a using the luciferase assay in Cal27 cells. The decreased luciferase activity was reported by miR-196a co transfection with pCheck2 reporter luciferase vector. Meanwhile, in order to clarify the target specificity, we established a mutant form of 3'-Untranslated Regions (3-UTRs) of ANXA1, where all 3 binding sites of miR-196a were destroyed by the Q5® Site- Directed Mutagenesis Kit (New England BioLabs Inc. (NEB)). The co-transfection of miR-196a and mutant vector did not change the luciferase activity, suggesting that ANXA1 was a direct target for miR- 196a (fig. 2A). The migration ability of Cal27 cells transfected with antagomiR-NC, antagomiR-196a or antagomiR-196a plus siAXAN1 was shown in fig. 2B. We used the antagomiR-196a technique to silence the expression activity of miR-196a. The wound healing assay indicated that repression of miR-196a significantly reduced the wound closure compared with antagomiR-NC, which was attenuated by cotransfection of ANXA1 siRNA (fig. 2B). The number of colonies measurements and invasion ability of Cal27 cells from different treatments was shown in fig. 2C and fig. 2D. The treatment of antagomiR-196a also decreased the number of colonies and inhibited cell invasion, which were all ANXA1 dependent (fig. 2C and fig. 2D).
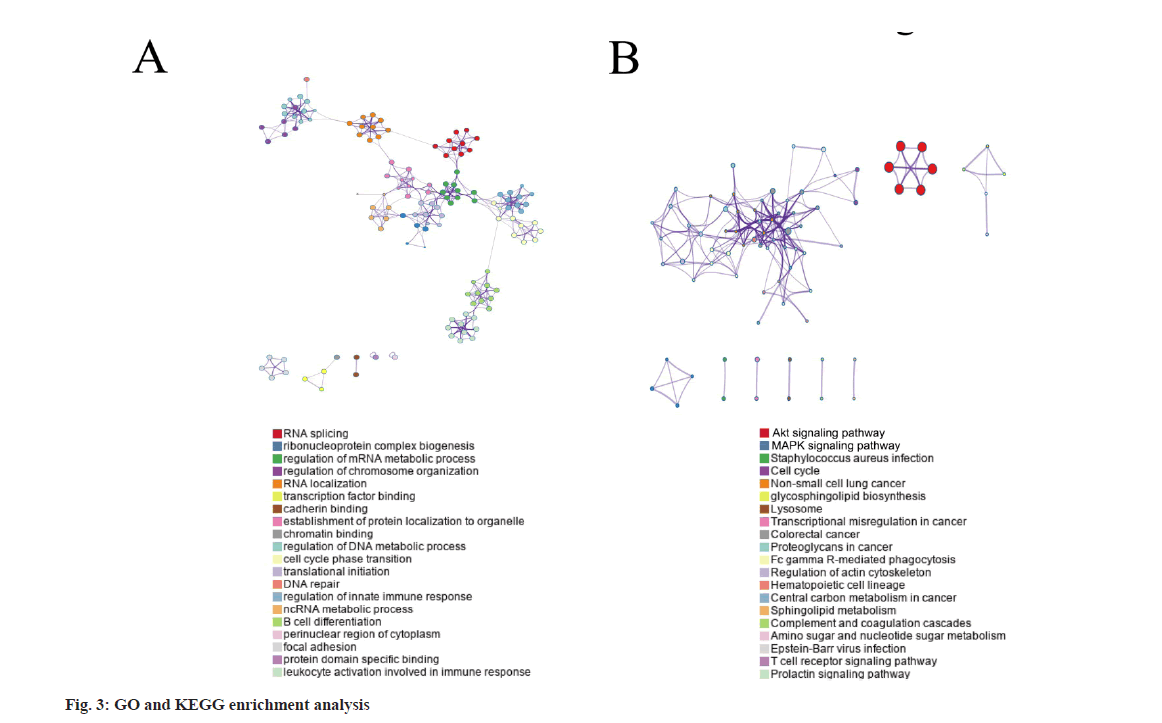
Results of GO and KEGG enrichment analysis was shown in fig. 3. The top GO term enrichment results with the largest number of genes were shown in fig. 3A. By performing GO and KEGG enrichment analysis on these DEGs, it could be found that the DEGs were enriched in GO terms including RNA splicing and ribonucleoprotein complex biogenesis (fig. 3A). Meanwhile, the Protein Kinase B (AKT) and Mitogen-Activated Protein Kinase (MAPK) signaling pathways were significantly enriched for KEGG analysis. The enrichment results of the KEGG pathways with the largest number of genes were shown in fig. 3B.
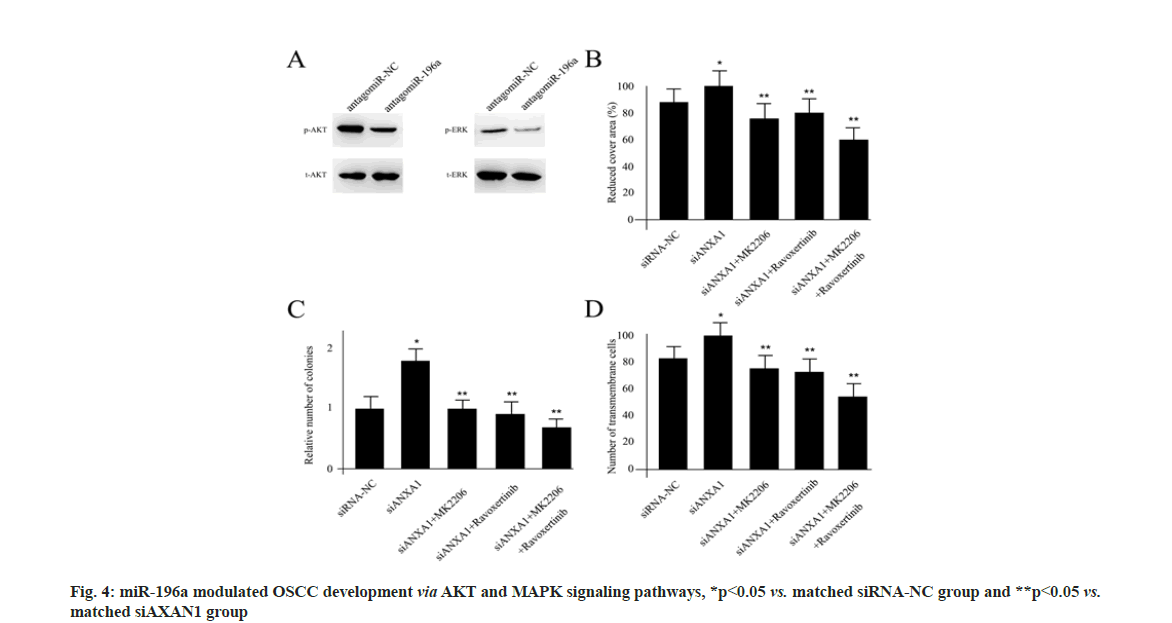
The miR-196a modulated OSCC development via AKT and MAPK signaling pathways. Since enrichment analysis suggested the AKT and MAPK signaling pathways as potential downstream regulators for miR-196a/ANXA1, we sought to verify the underlined molecular mechanism. In Cal27 cells, inhibiting the expression of miR-196a obviously blocked the activity of phosphorylated- AKT (p-AKT) and phosphorylated-Extracellular Signal-Regulated Kinase (p-ERK) using Western blotting assay (fig. 4A). The migration ability of Cal27 cells transfected with siRNA-NC, siAXAN1, siAXAN1+MK2206, siAXAN1+ravoxertinib and siAXAN1+MK2206+ravoxertinib was shown in fig. 4B. The 8 nM MK-2206 dihydrochloride (MK-2206) and 6.1 nM ravoxertinib were used as specific p-AKT and p-ERK inhibitor respectively. Based on the wound healing assay, transfection of ANXA1 siRNA in Cal27 cells significantly increased the wound closure compared with control, which was rescued by the treatment of MK-2206 and ravoxertinib individually or when combined together (fig. 4B). The number of colonies examination and invasion ability of Cal27 cells from different treatments was shown in fig. 4C and fig. 4D. The treatment of ANXA1 siRNA also enhanced the number of colonies and cell invasion, which were also repressed by MK-2206 and ravoxertinib administration (fig. 4C and fig. 4D).
The aggressiveness and complexity of OSCC have brought many obstacles to clinical treatment. The discovery of underlined molecular mechanism for OSCC patients is still a very strenuous journey. In just over two decades since the discovery of the first miRNA, the field of miRNA biology has expanded considerably. Accumulating insights into the functions of miRNAs in development and disease, particularly in cancer, have made miRNAs attractive tools and powerful targets for novel therapeutic approaches. Previously, Rajan and his colleagues established a miRNA based expression profiling using miRNA microarray in 30 tumors and 18 normal samples, which was further validated by qRT-PCR of patients tumor specimens[20]. A set of 19 most dysregulated miRNAs were demonstrated. At the same time, they confirmed that there was up-regulation of miR-196a, miR-21, miR-1237 and downregulation of miR-204. The miR-144 was associated with poor prognosis of OSCC patients whereas, the miR-196a/miR-204 expression ratio could be considered as best predictor for the disorder recurrence and OSCC patient survival. In this study, 23 miRNAs were suggested as potential DEMs for OSCC patients. The difference could be attributed to the variability of OSCC patient’s samples. However, the miR-196a was also considered as the primary DEMs from our report, which was consistent to the previous findings.
Based on the functional studies, researchers believe that miRNA dysregulation is the key causal factor in many cases of cancer, whereas miRNAs acting as either tumour suppressors or oncogenes (oncomiRs) based on distinct roles of miRNAs are also shown here. Moreover, the same miRNA might be functional and totally different in diverse organs. For example, the aberrant expression of miR-34 and its target genes in gastric cancer suggested its roles as a oncomiRs[21].
On the other hand, the downregulation of miR-34 family was shown to be a crucial part in ovarian cancer development and the low miR-34 levels were linked to a worse overall survival, indicating a tumor suppressor role for miR-34 in this respective[22]. The miR-196a was considered as oncomiRs by majority of the studies so far. A work by Lu et al. measured the stability of circulating miR-196a at different temperature conditions in plasma samples, including 53 from healthy individuals, 16 from pre-cancer patients and 90 from oral cancer patients[23]. They claimed that circulating miR-196a was very stable when storing plasma samples at -20° or below, but substantially increased in patients with oral precancer lesions (5.9 fold, p<0.01) and in oral cancer patients (9.3 fold, p<0.01), which suggested that the miR-196a could serve as a panel plasma biomarker for the early detection of OSCC.
Except for the potential DEMs, we also hypothesized several DEGs for OSCC patients, with ANXA1 as the most significant one. The ANXA1 stands for Annexin A1, which has become the hot spot for the current cancer research. For instance, overexpression of ANXA1 and Ephrin type-A receptor 2 (EphA2) has been shown to be closely associated with various cancers. ANXA1 could compete with Casitas B-Lineage Lymphoma (CBL) for binding EphA2 and enhance its stability by inhibiting CBL-mediated EphA2 ubiquitination as well as degradation in nasopharyngeal carcinoma. By these, ANXA1 was suggested to promote nasopharyngeal carcinoma cell growth and metastasis both in vitro and in vivo[24]. Moreover, in Human Epidermal Growth Factor Receptor-2 (HER-2+) breast cancer patients, ANXA1 was previously reported to contribute to trastuzumab resistance through AKT dependent signaling pathway[25]. The ANXA1 was considered as an oncogene in this process. However, the oncogenetic function was not unique for ANXA1 in the carcinogenesis. In prostate cancer, ANXA1 was one of the significant down-regulated DEGs[26]. The ANXA1 has been realized by many researchers for its multiple functions essential in cancer, including cell proliferation, apoptosis, chemosensitivity, metastasis as well as invasion. More importantly, ANXA1 expression was shown to vary depending on the tumor type and there were various contradictory reports on its role in the regulation proliferation and tumor growth[27].
Here, in this study, our data suggested that ANXA1 was functional as a tumor repressor for OSCC progression, which was also a key downstream regulator for miR-196a. With external experiments, it could be demonstrated that ANXA1 modulated OSCC cell migration, cell invasion and the number of colonies by inhibiting AKT and ERK phosphorylation, which could be rescued by MK- 2206 and ravoxertinib treatment individually or when combined together. As a calcium-dependent phospholipid binding protein identified in animals, plants as well as microorganisms, ANXA1 has been reported to participate in various cellular physiological activities. Using ANXA1 Knock Out (KO) and Over Expression (OE) mice, the laboratory group of Zhang conducted that ANXA1 could participate in hair growth by affecting the density of hair follicle through Epidermal Growth Factor (EGF) signaling pathway[28]. Similar to our findings, they also claimed that the expression of ANXA1 was negatively correlated with the levels of phosphorylated AKT1 and ERK2 proteins.
Collectively, in the light of fact that the molecular mechanism underlined OSCC is still poorly understood, we hypothesized a miR-196a and ANXA1 dependent signaling cassette for OSCC progression. Moreover, the downstream signaling pathway was also in-depth explored. All the integrated work here provided several new insights in the future OSCC basic and clinical study.
Conflict of interests:
The authors declared no conflict of interest.
References
- Chow LQ. Head and neck cancer. N Engl J Med 2020;382(1):60-72.
[Crossref] [Google scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google scholar] [PubMed]
- Leemans CR, Snijders PJ, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18(5):269-82.
[Crossref] [Google scholar] [PubMed]
- Zhuang RY, Xu HG. Head and neck cancer. N Engl J Med 2020;382(20):e57.
- Miranda-Filho A, Bray F. Global patterns and trends in cancers of the lip, tongue and mouth. Oral Oncol 2020;102:104551.
[Crossref] [Google scholar] [PubMed]
- Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol 2015;8(9):11884-94.
[Google scholar] [PubMed]
- Alves AM, Diel LF, Lamers ML. Macrophages and prognosis of oral squamous cell carcinoma: A systematic review. J Oral Pathol Med 2018;47(5):460-7.
[Crossref] [Google scholar] [PubMed]
- Chakraborty D, Natarajan C, Mukherjee A. Advances in oral cancer detection. Adv Clin Chem 2019;91:181-200.
[Crossref] [Google scholar] [PubMed]
- Sasahira T, Kirita T. Hallmarks of cancer-related newly prognostic factors of oral squamous cell carcinoma. Int J Mol Sci 2018;19(8):2413.
[Crossref] [Google scholar] [PubMed]
- Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci 2019;20(24):6249.
[Crossref] [Google scholar] [PubMed]
- Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: An overview. Methods Mol Biol 2017;1509:1-10.
[Crossref] [Google scholar] [PubMed]
- Wu M, Wang G, Tian W, Deng Y, Xu Y. MiRNA-based therapeutics for lung cancer. Curr Pharm Des 2017;23(39):5989-96.
[Crossref] [Google scholar] [PubMed]
- Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. Rna 2019;25(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets 2018;18(3):266-77.
[Crossref] [Google scholar] [PubMed]
- Guo Y, Qiao X, Zhu L, Song R. MicroRNA-182-5p modulates oral squamous cell carcinoma migration and invasion via targeting MTSS1 gene. Pathol Oncol Res 2020;26(2):1007-13.
[Crossref] [Google scholar] [PubMed]
- Zhang L, Liao Y, Tang L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res 2019;38(1):1-3.
[Crossref] [Google scholar] [PubMed]
- Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, et al. miRNA nanotherapeutics for cancer. Drug Discov Today 2017;22(2):424-32.
[Crossref] [Google scholar] [PubMed]
- Tan LP, Tan GW, Sivanesan VM, Goh SL, Ng XJ, Lim CS, et al. Systematic comparison of plasma EBV DNA, anti‐EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. Int J Cancer 2020;146(8):2336-47.
[Crossref] [Google scholar] [PubMed]
- Gao Y, Lin L, Li T, Yang J, Wei Y. The role of miRNA-223 in cancer: Function, diagnosis and therapy. Gene 2017;616:1-7.
[Crossref] [Google scholar] [PubMed]
- Rajan C, Roshan VG, Khan I, Manasa VG, Himal I, Kattoor J, et al. MiRNA expression profiling and emergence of new prognostic signature for oral squamous cell carcinoma. Sci Rep 2021;11(1):1-2.
[Crossref] [Google scholar] [PubMed]
- Jafari N, Abediankenari S. MicroRNA-34 dysregulation in gastric cancer and gastric cancer stem cell. Tumor Biol 2017;39(5):1010428317701652.
[Crossref] [Google scholar] [PubMed]
- Welponer H, Tsibulak I, Wieser V, Degasper C, Shivalingaiah G, Wenzel S, et al. The miR-34 family and its clinical significance in ovarian cancer. J Cancer 2020;11(6):1446-56.
[Crossref] [Google scholar] [PubMed]
- Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY, et al. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem 2015;48(3):115-21.
[Crossref] [Google scholar] [PubMed]
- Feng J, Lu SS, Xiao T, Huang W, Yi H, Zhu W, et al. ANXA1 binds and stabilizes EphA2 to promote nasopharyngeal carcinoma growth and metastasis ANXA1–EphA2 interaction and anticancer peptide. Cancer Res 2020;80(20):4386-98.
[Crossref] [Google scholar] [PubMed]
- Silva-Oliveira R, Pereira FF, Petronilho S, Martins AT, Lameirinhas A, Constâncio V, et al. Clinical significance of ARID1A and ANXA1 in HER-2 positive breast cancer. J Clin Med 2020;9(12):3911.
[Crossref] [Google scholar] [PubMed]
- Li D, Hao X, Song Y. Identification of the key MicroRNAs and the miRNA-mRNA regulatory pathways in prostate cancer by bioinformatics methods. Biomed Res Int 2018:1-10.
[Crossref] [Google scholar] [PubMed]
- Foo SL, Yap G, Cui J, Lim LH. Annexin-A1–a blessing or a curse in cancer? Trends Mol Med 2019;25(4):315-27.
[Crossref] [Google scholar] [PubMed]
- Zhang H, Fu X, Ao Y, Nan M, Qiu Z, Jia X, et al. ANXA1 affects murine hair follicle growth through EGF signaling pathway. Gene 2021;771:145343.
[Crossref] [Google scholar] [PubMed]