- *Corresponding Author:
- Y. Huo
Department of Anatomy, School of Basic Medicine, Hebei North University, Zhangjiakou, Hebei 075000, China
E-mail: yaocangdang5@163.com
| Date of Received | 24 July 2021 |
| Date of Revision | 14 May 2022 |
| Date of Acceptance | 13 September 2022 |
| Indian J Pharm Sci 2022;84(5):1279-1287 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the molecular pathway of the circular RNA molecule in gefitinib acquired resistance in nonsmall cell lung cancer and to provide a new therapeutic direction for clinical treatment of non-small cell lung cancer. The gefitinib-sensitive non-small cell lung cancer cell strain H292_S was selected to construct gefitinib acquired resistant cell model H292_R. Also, the overexpression group and vector control group of H292_S transfected circSETD3; the knockdown plasmid group and vector control group of H292_R transfected circSETD3 were prepared. The degree of cell response to gefitinib was measured by Western blotting and cell counting kit-8 method. The RNA sequencing method was used to screen differentially expressed circular RNA in cells and real-time fluorescent quantitative polymerase chain reaction was used to verify the sequencing results; the dual-luciferase report was used to screen and verify the relationship between circSETD3 and microRNA-520h. After the drug-resistant H292_R cell strain was processed by gefitinib, the expression of phosphorylated epidermal growth factor receptor, phosphatidylinositol 3-kinase and protein kinase B decreased significantly. The expressions of three circular RNAs in has_circ_0000567, has_circ_0000655 and has_circ_0006867, were all up-regulated in drug-resistant strains, and the differences were statistically significant (p<0.05). The overexpression group of circSETD3 had a significant effect on the growth and reproduction ability of cells (p<0.05); the wild type group of the dual-luciferase report plasmid was transfected into 293T cells together with microRNA-520h mimics and the fluorescence intensity decreased significantly (p<0.05). In addition, if the luciferase reporter plasmid mutant group (mutant) was transfected into 293T cells together with microRNA-520h mimics, the fluorescence intensity would not change significantly. circSETD3 could be directly bound to microRNA-520h.
Keywords
circSETD3, non-small cell lung cancer, gefitinib, drug resistance, tyrosine kinase
Among the tumors with the highest incidence in the world, Non-Small Cell Lung Cancer (NSCLC) accounts for more than 80 %[1,2]. The disease is not easily detected in the early stage; so many patients are discovered in the middle or late stages. At this time, conventional radiotherapy and chemotherapy are not very effective and the survival rate of patients within 5 y is only about 3/20[3]. Based on this condition, a new molecularly targeted drug gefitinib for the treatment of NSCLC has received extensive attention from the medical community and has also made great progress.
Gefitinib is a kind of tyrosine kinase inhibitor, which can compete for adenine nucleoside triphosphate and bind to extracellular ligand sites, thereby blocking the activation process of intracellular tyrosine kinase and ultimately interrupting cell growth[4,5]. Gefitinib has small side effects, so it has been widely used in clinical treatment in the medical field. However, many patients will develop acquired resistance after using gefitinib for more than 6 mo[6]. Current researches show that the emergence of gefitinib resistance may be related to the overexpression of the Adenosine Triphosphate (ATP)-Binding Cassette (ABC) protein family of efflux transporters. The specific molecular mechanism is not clear. Circular RNA (circRNA) is endogenous non-coding RNAs that do not contain 3’ and 5’ ends. They are RNA molecules with a closed circular structure.
The molecular characteristics of circRNAs are not easy to be decomposed by RNAse, with strong stability, highly conservative and disease-specific[7,8]. CircRNAs have an important relationship with the occurrence and prognosis of a variety of malignant tumors and are potential clinical intervention targets[9]. However, whether circRNAs affect the sensitivity of targeted drugs such as gefitinib in tumor cells has not been reported in the literature. Only reports indicate that circRNAs can activate the epidermal growth factor signaling pathway in tumor cells. And epidermal growth factor is the specific inhibitory target of gefitinib; the reforests can be speculated that circRNAs are likely to induce drug resistance of tumor cells during the administration of gefitinib.
In response to the above problems, a NSCLC cell line model of gefitinib acquired resistance was established and RNA sequencing technology was employed to screen out the cyclic RNA (circSETD3) with the most significant difference in expression before and after drug resistance. The molecular mechanism of circSETD3 in the process of acquiring gefitinib resistance was analyzed, to provide experimental evidence for the precise treatment of clinical NSCLC.
Materials and Methods
Materials:
NSCLC cell line H292 (Chinese Academy of Medical Sciences).
Cell culture:
Resuscitation: Complete medium was configured in a ratio of medium/serum=9/1 and 1:100 double antibodies (penicillin and streptomycin) (Tiangen Biotech (Beijing) Co., Ltd., China) were added. 5 ml of complete medium was added to an empty centrifuge tube (Tiangen Biotech (Beijing) Co., Ltd., China) and the thawed cells were poured into the medium. The mixture was centrifuged at 1200 rpm for 3 min and the supernatant was discarded. Then, 4 ml of complete medium was added and blended well. After that, it was placed in a cell incubator containing 5 % Carbon dioxide (CO2) and incubated overnight at 37°. Attention was paid to the adherence of the cells and the medium was supposed to be replaced in time.
Medium change: The culture medium where there were no cells were discarded and the culture medium was washed with Phosphate-Buffered Saline (PBS) (Tiangen Biotech (Beijing) Co., Ltd., China) for three times. Then, the PBS was discarded and 5 ml of complete medium was added. The medium was incubated in a cell incubator containing 5 % CO2, at 37°.
Cell passage: The complete medium with a ratio of medium/serum=9/1 was configured and 1:100 double antibodies (penicillin and streptomycin) were added. The cells were observed under a microscope (Leica Company, Germany). When the confluence rate exceeded 70 %, the culture solution was poured out and the medium was rinsed with PBS 3 times. Then, 1 ml of 0.25 % pancreatin (Tiangen Biochemical Technology (Beijing) Co., Ltd., China) was added and the cells were put in an incubator for digestion. After the cells were detached, 2 ml of complete medium was added at the ratio of trypsin:complete medium is equal to 1:2 to terminate the digestion. After the cells were in a single floating state through continuous blowing, the cell suspension was transferred to a centrifuge tube and centrifuged at 1000 rpm for 8 min. The supernatant was discarded and complete medium was added to mix. 1 ml of cell suspension was put into the 3 ml of a new complete medium. The solution was mixed well and put into the incubator.
Cryopreservation: To subculture the cells according to the supernatant after centrifugation was discarded. 1 ml frozen stock solution was added. After the mixture was blended evenly, it was transferred to the cryopreservation tube and placed in a refrigerator (Haier Group, China) at 4° for 30 min first, then in the refrigerator at -20° (Thermo, United State of America (USA)) for 2 h and finally, in the refrigerator at -80° (Thermo Company, USA). After a whole night, the cells were stored in liquid nitrogen.
The establishment of gefitinib acquired resistant in NSCLC cell line model:
The NSCLC cell H292 was placed in a cell incubator (Shanghai Yiheng Experimental Company, China) containing 5 % CO2 at 37°. 1 % double antibodies (penicillin and streptomycin) and 12 % fetal bovine serum (BI Company, Israel) were added into Roswell Park Memorial Institute (RPMI) 1640 and the gefitinib (AstraZeneca, United Kingdom) at low to high concentrations were added to the cell culture medium within 4 d. Then, the cells were digested with 0.25 % trypsin and passaged. Also, the cell growth status was observed at any time. 1 y later, the cells grew stably in the complete medium containing 2.0 μmol/l gefitinib. Gefitinib was removed 6 mo before the test to prevent the test results from being affected.
Cell proliferation detection:
Cells in the logarithmic growth phase after digestion and centrifugation were inoculated into a 96-well plate (Tiangen Biotech (Beijing) Co., Ltd., China) at a concentration of 40 000 cells/ml per well. 6 replicates were set per well and 100 μl PBS was added to the well. The 96-well plate was placed in a 5 % CO2 cell incubator and incubated at 37° for 48 h. 10 μl Cell Counting Kit- 8 (CCK-8) reagents to be added to each well. It was required no bubbles during the operation. And it was put into the cell culture box until the color changes. The micro plate reader (Biotek, USA) was used to measure the absorbance of each well.
Cell viability=(Experimental well-blank well)/(control well-blank well)×100 %.
Western blotting detection:
Extract total protein: The cells were taken out and the culture solution was discarded. After that the culture medium was rinsed with PBS, 70 μl cell lysate was added to each well. In 5 min, the cell suspension was transferred to an Eppendorf (EP) tube (Tiangen Biotech (Beijing) Co., Ltd., China). The tube was shaken once every 5 min for a total of 6 times. After that it was put in a 4° centrifuge, to centrifuge at 1000 rpm for 15 min. The supernatant was taken for Bicinchoninic Acid (BCA) protein quantitative determination and the standard curve was drawn.
The stacking gel and separation gel were prepared and the reagents (Tiangen Biotech (Beijing) Co., Ltd., China) were shown in Table 1 below.
| Elements | The stacking gel | 10 % separation gel |
|---|---|---|
| Double distilled water | 2.7×103 | 4×103 |
| 30 % Polyacrylamide solution | 0.67×103 | 3.3×103 |
| 1.0 mol/l Tris (tromethamine, pH 8.8) | 2.5×103 | 2.5×103 |
| 10 % Sodium dodecyl sulfate | 0.04×103 | 0.1×103 |
| 10 % Ammonium persulfate | 0.041×103 | 0.1×103 |
| Tetramethylethylenediamine | 0.004×103 | 0.004×103 |
Table 1: Configuration of Stacking Gel and Separation Gel
Electrophoresis and development: After the glass plates were thoroughly cleaned by distilled water and ethanol, the glass plates were aligned and tightly clamped. Then, they were vertically stuck on the glue rack. Distilled water was added to the glass plates to a suitable position and through 8 min of standing, it could be seen that whether the glass plates leaked or not. 10 % of separation gel was prepared according to the formula in Table 1. After mixing, 6 ml was added to the gap between the glass plates with a pipette and then 3 ml of isopropanol was added slowly. Then, the plates were placed at 37° until a refraction line showed up between the isopropanol and the separated gel which meant the separated gel solidified. The isopropanol was discarded and the glass plates were washed with distilled water three times for later use. 3 ml stacking gel was added to the glass plates after it was configured and the comb was slowly placed inside without producing bubbles.
After the stacking gel was solidified, the glass plates and plastic replacement plates were clamped into the rack with electrodes and put into the electrophoresis tank. Meanwhile, the comb was pulled out. 30 μl of the expressed protein supernatant was taken and 10 μl of 5× loading buffer was added and mixed well. Then the mixed solution was boiled for 10 min at 100°.
Finally, 40 μl electrophoresis gels were applied to each well. At 80V voltage, the bromophenol blue formed a straight line in the gel and then the voltage was changed to 120V. When the bromophenol blue went to the lower edge, the power supply was disconnected and the membrane was transferred. After the transfer was completed, the gel image processing system (Unverbindlicher Verkaufspreis, Germany) was employed to analyze the molecular weight and net Optical Density (OD) of the objective.
The relative expression of the target protein=target band grey value OD/internal reference grey value OD
Differential circRNAs detection by RNA sequencing and real-time fluorescent quantitative Polymerase Chain Reaction (PCR):
RNA sequencing: HiSeqTM 2500 sequencer (Illumina Company, USA) was used to perform RNA sequencing on H292_S and H292_R. First, a database was built for RNA whose standard was RNA Integrity Number (RIN) ≥7, 28S/18S>0.7. The circRNAs with a Fold Change (FC) ≥2 and p<0.05 during the sequencing process could be referred to as circRNAs that were differentially expressed in sensitive cells and drugresistant cells.
The RNA extraction kit (Tiangen Biotech (Beijing) Co., Ltd., China) was used to extract total RNA and then complementary DNA (cDNA) was obtained in the condition of 37° for 15 min and 85° for 5 s according to the reverse transcription kit (Tiangen Biotech (Beijing) Co., Ltd., China). The obtained cDNA was stored at -80°.
Fluorescence quantitative PCR (Tiangen Biotech (Beijing) Co., Ltd., China): First, total RNA was extracted, after the medium was rinsed with PBS, 1 ml of Trizol reagent was added. After they were mixed and stood at room temperature for 15 min, the cell lysate and 200 μl of chloroform were added to the EP tube. Then, they were shaken for 10 s and stood at room temperature for 5 min. Later, the mixture was centrifuged for 10 min under 13 000 rpm at 4°. The supernatant (RNA) was carefully transferred to a new EP tube and the volume was recorded. As soon as the same volume of isopropanol was added, the solution was shaken for 10 s; then, with 10 min standing on ice, it was centrifuged at 13 000 rpm at 4° for 10 min. The obtained supernatant was carefully extracted, 1 ml of 75 % ethanol solution was added, shaken, mixed and centrifuged at 13 000 rpm under 4° for 4 min. Moreover, after the mixture was placed at room temperature for 30 min, 20 ml Diethyl Pyro Carbonate (DEPC) was added. The OD260/OD280 value of RNA was measured and then the mixed solution was sub packaged and put into -80° refrigerator.
After reverse transcription of RNA, fluorescence quantitative PCR was performed in the various reaction conditions of 95°, 30 s; 95°, 5 s; 60°, 30 s; for 39 cycles. The reaction reagents were shown in Table 2.
| Reagent | SYBR premix enzyme | Forward primer (10 µM) | cDNA | DEPC |
|---|---|---|---|---|
| Dose | 5 µl | 0.5 µl | 0.5 µl | 3 µl |
Table 2: Fluorescence Quantitative PCR System
Effect of circSETD3 on cell gefitinib sensitivity:
The overexpression plasmid and interference plasmid of circSETD3 were transfected into sensitive and drug-resistant strains and the transfection efficiency was verified by real-time fluorescent quantitative PCR. After gefitinib was added, the CCK-8 method and flow cytometry were used to detect cell growth and apoptosis.
Cell transfection: After cell digestion, 1.0 μg circSETD3 overexpression plasmid was diluted with 200 μl serum-free RPMI 1640 medium (Gibco, USA) and then 200 μl serum-free RPMI 1640 medium was taken to dilute 10 μl LipofectamineTM 2000 transfection reagent (Gibco, USA). After 8 min standing and being mixed gently, the two reacted for 15 min at room temperature. 2 ml of serum-free RPMI 1640 medium was added to a 96-well plate and then the mixture was added drop wise. The plate was placed in a cell culture incubator containing 5 % CO2 and incubated at 37° for 1-3 d (cell growth was observed to replace the complete medium).
Detection of cell apoptosis: The cells in the logarithmic growth phase were inoculated into a 6-well plate with 1×105 cells/ml, 2 ml per well and placed in a cell incubator containing 5 % CO2 and cultured at 37°. When the cells confluence rate exceeded >80 %, the circSETD3 overexpression plasmid and the control vector plasmid were transfected into the cells. After 5 h, the medium was rinsed 3 times with PBS and replaced with complete culture medium. The cells were cultured in the same conditions for another 3 d. After digestion with trypsin, the medium was centrifuged for 18 min and the supernatant was discarded. In addition, PBS was added to mix well, 100 μl binding buffer was added and transferred to flow detection tube. With the addition of 5 μl Annexin V (Tiangen Biotech (Beijing) Co., Ltd., China), they reacted at room temperature under shaking for 18 min. After 15 min centrifugation at 1000 rpm, the staining solution was discarded. At last, 100 μl of binding buffer was added and mixed, then 5 μl of Propidium Iodide (PI) staining solution (Tiangen Biotech (Beijing) Co., Ltd., China) was added and mixed well under shading. The solution was filtered with a 0.45 μm filter, the filtered liquid was stored on ice and protected from light. The sample was analyzed through a FACSCaliburTM flow cytometer (BD Company, USA).
Dual-luciferase report gene assay:
Cell transfection: The transfection reagent was prepared and the test was started when the cell concentration was around 60 %. 10 μl (Dulbecco’s Modified Eagle Medium (DMEM), BGI, China) was mixed with 0.14 μg circSTED3/mutant and circSTED3/Wild-Type (circSTED3/WT) target plasmid and then 5 pmol of micro RNA (miR)-520h/negative. Control (BGI, China) was added and mixed well and stood at room temperature (solution M). After that 1 μl DMEM was mixed with 0.3 μl of transfection reagent thoroughly (solution N) and stood at room temperature for 10 min. The solution M and solution N was put together and stood at room temperature. The culture medium was replaced after 6 h and cells were tested after 2 d.
100 μl 1× Passive Lysis Buffer (PLB, BGI, China) was added to each well of a 96-well plate and placed on a shaker at room temperature. After being shaken slowly for 20 min, the cell lysate was transferred to a 1.5 ml centrifuge tube and centrifuged for 10 min at 12 000 rpm, 4°. The supernatant was transferred to a new tube. Then 100 μl Luciferase Assay Reagent II (LAR, BGI, China) and 20 μl cell lysate were added to the 96-well plate. The value of firefly luciferase was measured and recorded as an internal reference value. At last, 100 μl Stop and Glo Reagent (BGI Gene, China) was added and Renilla luciferase was measured (to report gene luminescence value) after mixing.
Statistical analysis:
Statistical Package for the Social Sciences (SPSS) 26.0 statistical software was applied to perform statistical difference analysis of the results and the measurement data was analyzed by the equation; mean±standard deviation (x±s) and independent sample t-test. Moreover, the chi-square test was performed for enumeration data. If p<0.05, the difference was considered statistically significant.
Results and Discussion
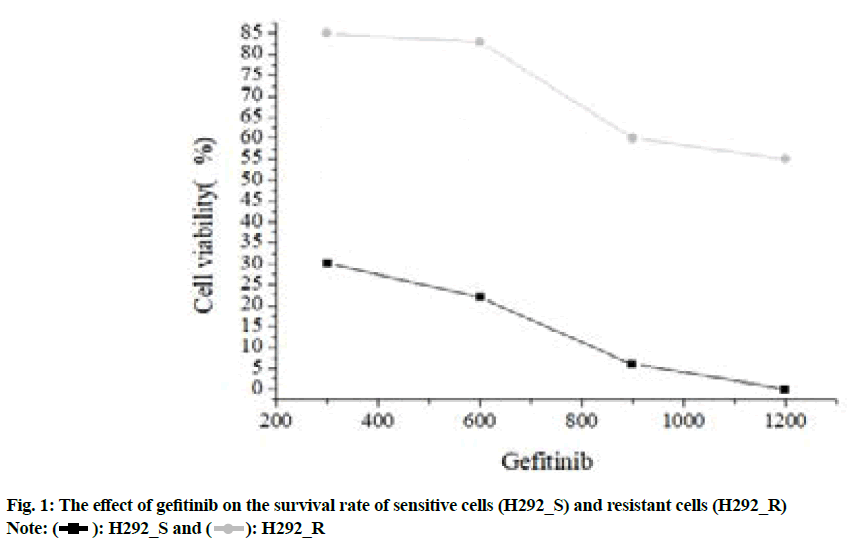
The CCK-8 method was used to detect gefitinib acquired resistant NSCLC cell lines. The results were shown in fig. 1.
Fig. 1 showed the inhibitory concentration of gefitinib on the growth and proliferation of H292_S and H292_R cells were 107.58 nmol/l and 979.27 nmol/l respectively and the drug resistance index was 9.01.
Western blotting was used to detect the effect of gefitinib on the signaling pathway of drug-resistant cells, as shown in fig. 2.
Fig. 2 indicated after treating the drug-resistant H292_R cells with 2 μmol/l gefitinib, the expression of phosphorylated Epidermal Growth Factor Receptor (p-EGFR), phosphorylated Phosphatidylinositol 3-Kinase (p-PI3K) and phosphorylated Protein kinase B (p-AKT) decreased significantly and the above three were the activation forms of the EGFR pathway, which could indicate that in the drug-resistant cell H292_R, gefitinib treatment reduced the inhibitory function of the EGFR signaling pathway, therefore, the sensitivity of the cells to gefitinib was significantly reduced.
The total RNA of gefitinib acquired resistant NSCLC cell strains were subjected to high-throughput sequencing (BGI Gene, China). The sequencing results were shown in Table 3.
| circRNA ID | hsa_circ_0000567 | hsa_circ_0000655 | hsa_circ_0000722 | hsa_circ_0006867 |
|---|---|---|---|---|
| Gene symbol | SETD3 | CHD2 | GSE1 | NAA15 |
| circRNA_chr | NC_000014.9 | NC_000015.10 | NC_000016.10 | NC_000004.12 |
| circRNA_length/bp | 683 | 413 | 219 | 263 |
Table 3: Differentially Expressed circRNA
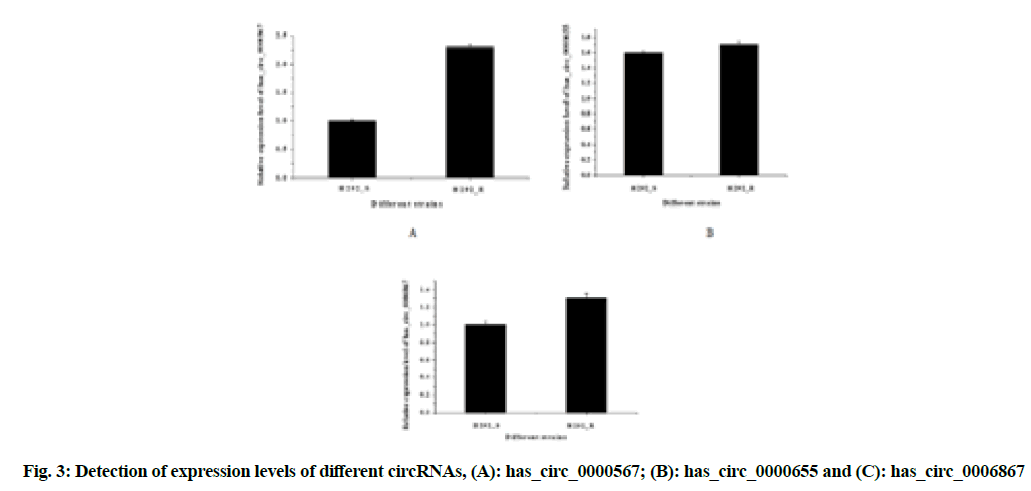
In order to verify the accuracy of RNA-sequencing high-throughput sequencing results, the real-time fluorescent quantitative PCR was used to verify the expression of three up-regulated circRNAs in drug-resistant strains, as shown in fig. 3A-fig. 3C.
The expression of the three circular RNAs, has_circ_0000567, has_circ_0000655 and has_ circ_0006867, were all up-regulated in drug-resistant strains and the difference was statistically significant (p<0.05); while the difference of has_circ_0000567 (circSETD3) was the most obvious, which could be studied from the perspective of molecular mechanism.
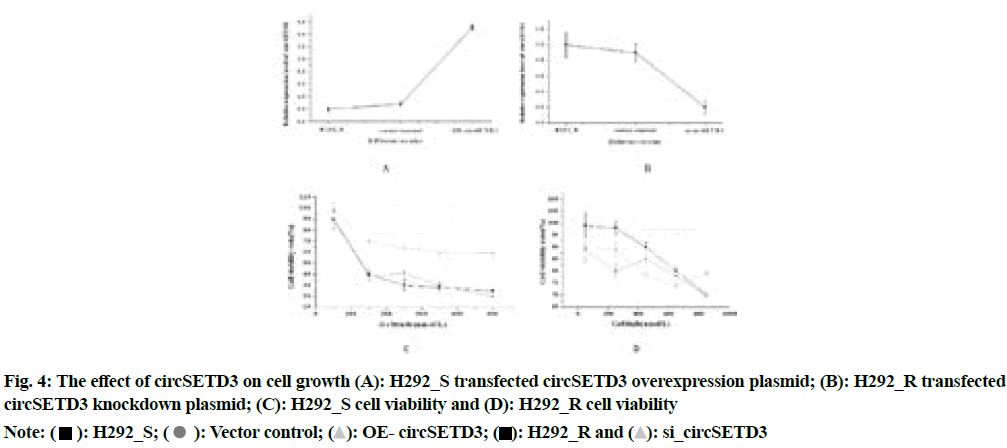
The circSETD3 overexpression plasmid and its negative control were transfected into H292_S and the circSETD3 knockdown plasmid and its negative control were transfected into H292_R. The detection result of CCK-8 was shown in fig. 4A-fig. 4D.
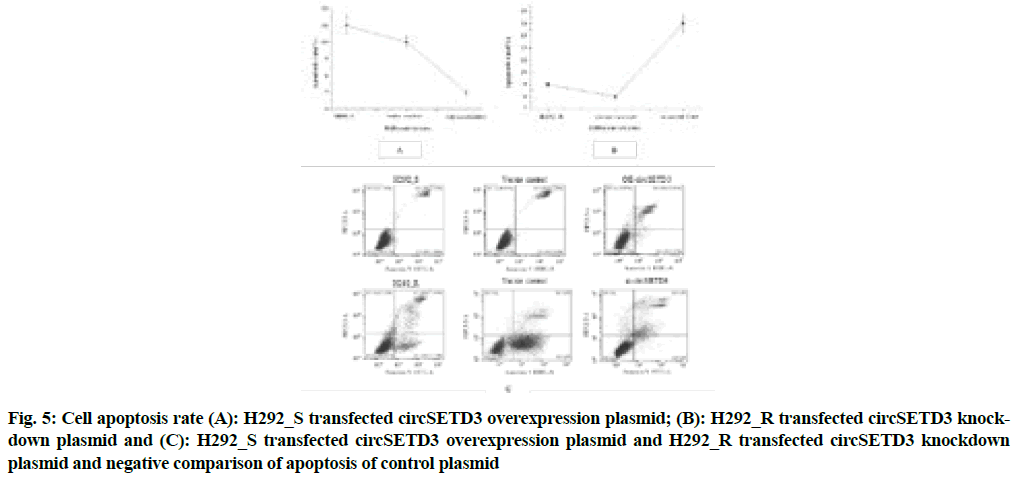
Fig. 4 above illustrated that compared with the negative control group, the overexpression group of circSETD3 had a very strong influence on the growth and proliferation of cells (p<0.05), the significant reduction of cell proliferation ability of the circSETD3 knockdown group (p<0.05), indicated that circSETD3 could promote cell growth and proliferation. The results of flow cytometry were shown in fig. 5A-fig. 5C.
Fig. 5: Cell apoptosis rate (A): H292_S transfected circSETD3 overexpression plasmid; (B): H292_R transfected circSETD3 knockdown plasmid and (C): H292_S transfected circSETD3 overexpression plasmid and H292_R transfected circSETD3 knockdown plasmid and negative comparison of apoptosis of control plasmid
Fig. 5 showed that compared with the negative control group, the apoptotic rate of the circSETD3 overexpression group decreased significantly (p<0.05), indicating that circSETD3 could inhibit cell apoptosis.
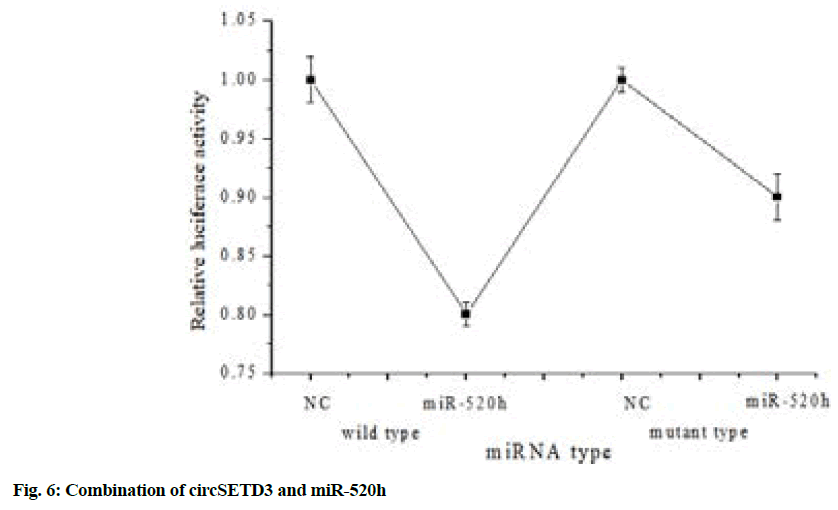
The constructed dual-luciferase reporter plasmid was transfected into 293T cells together with miR-520h mimics and the result of fluorescence intensity detection was shown in fig. 6.
Fig. 6 above illustrated that the WT of the dualluciferase reporter plasmid was transfected into 293T cells together with miR-520 h mimics and the fluorescence intensity decreased significantly (p<0.05). But when the luciferase reporter plasmid mutant group (mutant) was transfected into 293T cells together with miR-520h mimics, the fluorescence intensity did not change significantly, which showed that circSETD3 could directly bound to miR-520h.
RNA sequencing technology is used to detect differentially expressed circRNAs during the process of gefitinib acquired resistance. Compared with the traditional gene chip technology, this technology is characterized by high throughput, high sensitivity, fast sequencing speed, good repeatability and a wide range of applications[10]. At the same time, RNA sequencing technology can sequence the entire genome and can efficiently discover new genes, unknown transcripts or spliceosomes, whose result is also equivalent to the result of real-time fluorescent quantitative PCR, therefore, it is currently a hotspot in the field of the transcriptase[11]. CircRNAs generally have different locations and can produce different effects. The circRNA located in the nucleus generally interacts with DNA or protein, thereby affecting the expression of target genes. According to Du et al.[12], circRNAcircFOXO3 can interact with cyclin-dependent kinases to form protein complexes, which have an effect on the development of cell cycles and ultimately inhibit cell growth.
By consulting related literature, it is found that circSETD3-miR-520h-ATP-Binding Cassette subfamily G member 2 (ABCG2) can affect each other. The interaction between circSETD3 and miR-520h is confirmed by fluorescence quantitative PCR, dualluciferase report experiment and Western blotting. The discovery of Wang et al.[13] suggests that the overexpression plasmid transfected miR-520h can detect the decrease in expression of endogenous ABCG2, which may be because miR-520h inhibits translation or causes the instability of miR, thereby down-regulating ABCG2 expression. This is highly consistent with the results of the experiments and helps to further confirm the interaction between circSETD3- miR-520h-ABCG2.
It was confirmed that circSETD3 could promote tumor cell proliferation and inhibit apoptosis. Highly expressed circSETD3 could bind to miR-520h, which was more likely to promote the expression and function of ABCG2, which would eventually lead to the occurrence of acquired resistance to gefitinib. The result was conducive to the study of the factors affecting the secondary resistance of gefitinib and provided a strong basis for the treatment and prognosis of patients with clinical NSCLC.
RNA-sequencing was used to screen the circRNA whose expression was significantly up-regulated in gefitinib acquired resistance in NSCLC as circSETD3. Through light quantitative PCR, Western blotting and dual-luciferase report, it was confirmed that circSETD3 could promote the growth of lung cancer cells and inhibit cell apoptosis. CircSETD3 together with a huge amount of miR-520h may participate in the expression of ABCG2, which led to the emergence of acquired resistance to gefitinib. There are certain short comings, because of the lack of time, the expression of ABCG2 under this realization condition were not verified, which would also be part of the next experiment.
Conflict of interests:
The authors declared no conflict of interests.
References
- Park JS, Kim HR, Hong MH, Cho BC. P2. 06-003 A phase Ib study of the combination of afatinib and ruxolitinib in EGFR mutant non-small cell lung cancer (NSCLC) progressed on EGFR-TKI: Topic: Phase I trials. J Thorac Oncol 2017;12(1):S1069-70.
- Pietras R, Xu H, Hu X, Matheny C, Sandler A, Patel M. P1. 04-33 retrospective descriptive analysis of metformin with atezolizumab in advanced non-small cell lung cancer in the OAK trial. J Thorac Oncol 2018;13(10):S538-9.
- Patil T, Smith D, Bunn P, Aisner D, Le A, Hancock M, et al. P1. 01-78 the incidence of brain metastases in ROS1-rearranged non-small cell lung cancer at diagnosis and following progression on crizotinib. J Thorac Oncol 2018;13(11):1717-26.
[Crossref] [Google Scholar] [PubMed]
- Simasi J, Oelkrug C, Schubert A, Nieber K, Gillissen A. The role of BIM-EL and BCL2-α on the efficacy of erlotinib and gefitinib in lung cancer. Respir Physiol Neurobiol 2015;209:64-8.
[Crossref] [Google Scholar] [PubMed]
- Soria JC, Wu YL, Nakagawa K, Kim SW, Yang JJ, Ahn MJ, et al. Gefitinib plus chemotherapy vs. placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): A phase 3 randomised trial. Lancet Oncol 2015;16(8):990-8.
[Crossref] [Google Scholar] [PubMed]
- Maeda M, Murakami Y, Watari K, Kuwano M, Izumi H, Ono M. CpG hypermethylation contributes to decreased expression of PTEN during acquired resistance to gefitinib in human lung cancer cell lines. Lung Cancer 2015;87(3):265-71.
[Crossref] [Google Scholar] [PubMed]
- Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer. Oncol Rep 2015;33(6):2669-74.
[Crossref] [Google Scholar] [PubMed]
- Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids 2019;16:118-29.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Yang Y, Wang Z, Fu X, Chu XM, Li Y, et al. Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis 2020;298:14-26.
[Crossref] [Google Scholar] [PubMed]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell 2015;58(4):610-20.
[Crossref] [Google Scholar] [PubMed]
- Salleh MS, Mazzoni G, Höglund JK, Olijhoek DW, Lund P, Løvendahl P, et al. RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high-and low-residual feed intake in Nordic dairy cattle. BMC Genomics 2017;18(1):258.
[Crossref] [Google Scholar] [PubMed]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44(6):2846-58.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Xue Y, Wang P, Zhu J, Ma J. MiR-608 inhibits the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor. Oncol Rep 2016;35(5):2733-42.
[Crossref] [Google Scholar] [PubMed]
 H292_R
H292_R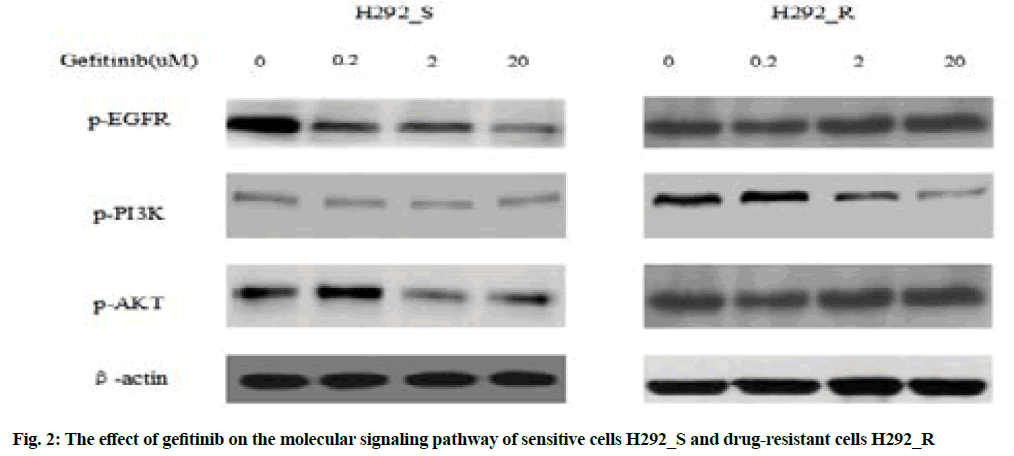
 Vector control;
Vector control;  OE- circSETD3;
OE- circSETD3;  si_circSETD3
si_circSETD3