- *Corresponding Author:
- S. Q. Li
Henan Center for Engineering and Technology Research on Prevention and treatment of Liver Diseases, Luoyang 471003, China
E-mail: sanqiangli2001@163.com
| Date of Received | 18 November 2021 |
| Date of Revision | 02 June 2022 |
| Date of Acceptance | 08 December 2022 |
| Indian J Pharm Sci 2022;84(6):1566-1575 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A disintegrin and metalloprotease 9 has been reported to play a key role in many diseases. However, the underlying effects and mechanisms of a disintegrin and metalloprotease 9 in alcoholic liver fibrosis have not been well understood. To investigate the role of a disintegrin and metalloprotease 9 and its regulatory molecular mechanism in alcoholic liver fibrosis, a single guide ribonucleic acid targeting the a disintegrin and metalloprotease 9 sequence of mice was adopted by clustered regularly interspaced short palindromic repeats/Cas9 technology to inhibit its expression in this study. Subsequently, hepatic stellate cell-T6 and male mice C57BL/6J were respectively divided into 4 groups and each group was treated differently. In vivo experiment, the normal group was fed with control Lieber-DeCarli TP4030C; the saline+alcohol group was injected with saline; the A disintegrin and metalloprotease 9-single guide ribonucleic acid+alcohol group was injected with effective single guide ribonucleic acid plasmids and the c-Jun N-terminal kinase inhibitor+alcohol group was injected with c-Jun N-terminal kinase inhibitor SP600125. All mice except for the normal group received Lieber-DeCarli TP4030A and carbon tetrachloride. Furthermore, examination of serum aspartate transaminase and alanine transaminase activities and the pathological analysis by hematoxylin-eosin staining, picric acid-sirius red staining, Hoechst 33 258 staining confirmed the pro-fibro genic effect of a disintegrin and metalloprotease 9. Compared with the saline+alcohol group, the levels and the expression of a disintegrin and metalloprotease 9, p-c-Jun, cytochrome P450 2E1, tumor necrosis factor alpha, alpha-smooth-muscle actin and Bcl-2-associated X in the A disintegrin and metalloprotease 9- single guide ribonucleic acid+alcohol group and the c-Jun N-terminal kinase inhibitor+alcohol group decreased significantly (p<0.05 or p<0.01), while the expression of heat shock protein 70, proliferating cell nuclear antigen and vascular endothelial growth factor increased significantly (p<0.05 or p<0.01). In addition, the disintegrin and metalloprotease 9-single guide ribonucleic acid+alcohol group and the c-Jun N-terminal kinase inhibitor+alcohol group in vitro experiment could inhibit the proliferation of hepatic stellate cells and promote apoptosis. Collectively, our results indicated that a disintegrin and metalloprotease 9 could promote alcoholic liver fibrosis in mice by activating the c-Jun N-terminal kinase signaling pathway, which is expected to make a breakthrough in the gene therapy of alcoholic liver fibrosis.
Keywords
A disintegrin and metalloprotease 9, alcoholic liver fibrosis, hepatic stellate cells, molecular mechanism
Alcoholic Liver Fibrosis (ALF), the most common Alcoholic Liver Disease (ALD), plays a key role in the later development of liver cirrhosis or liver cancer and the prognosis of the disease, which needs to be explored further in target therapy[1]. Research shows that about 50 % of patients who die of liver cirrhosis are caused by ALF[2]. In the process of liver fibrosis, alcohol can induce Hepatic Stellate Cells (HSC) activation[3] and massive acetaldehyde accumulating in the liver causes the synthesis of collagen I, which leads to Extracellular Matrix (ECM) deposition and liver tissue structural remodelling[4]. In recent years, it has been discovered that long-chain non-coding Ribonucleic acid (RNA) and microRNA (mRNA) are also involved in the activation of HSC[5,6]. The progress of ALF is also closely related to obesity, viral liver disease, genetic polymorphism, heredity, smoking and gender[7].
A Disintegrin and Metalloprotease 9 (ADAM9) plays an important role in various activities such as ECM degradation, cell adhesion, signal transduction and cell fusion and is involved in the formation, invasion, metastasis of various tumours in vivo[8-10]. Schwettmann et al.[10] found that the level of ADAM9, mRNA expression in activated HSC of chronic liver diseases caused by different etiologies was significantly higher than that of quiescent stellate cells and deduced that ADAM9 in the process of liver fibrosis disease may play an important role. Recently, it has been shown that ADAM9 has a function in promoting liver injury during an acute alcoholic liver injury in mice[11]. However, the impact of ADAM9 on ALF has not been well studied.
C-Jun N-Terminal Kinase (JNK) is known as the Mitogen-Activated Protein Kinase (MAPK) family[12], closely related to the occurrence and development of malignant tumours, which is mainly involved in cell growth, apoptosis, invasion, proliferation, metabolism and Deoxyribonucleic Acid (DNA) damage repair[13]. Previous studies have found that JNK signalling pathway is activated in a variety of liver stress and injury[14]. Qiu
[15] showed that the application of JNK inhibitor SP600125 can specifically block the JNK signal transduction pathway and protect the liver from ischemia-reperfusion injury, suggesting that JNK can be used as a target to treat diseases. At present, there are few reports on the activation of the JNK signalling pathway and the mechanism of ALF. It hopes that this study can provide new ideas and methods for clinical treatment.
This study was undertaken by using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 technology for targeted silencing of the ADAM9 gene. Through a series of experiments in vivo and in vitro, pathological change and the expression of related factors were detected to explore the role and molecular regulatory mechanism of ADAM9 in ALF, aiming to bring new ideas and approaches for clinical gene therapy of ALF.
Materials and Methods
Construction of plasmid targeting mutant mouse ADAM9 gene:
Firstly, the sequence of ADAM9 related genes was searched in National Centre for Biotechnology Information (NCBI) and the CRISPR sequence targeting mutation (single guide Ribonucleic Acid (sgRNA) sequence) was designed at exon 11. Then a three-in-one recombinant plasmid was constructed which expressed sgRNA, Cas9 and puromycin screening markers at the same time (provided by Nanjing YSY Biotech Ltd, China). At last, the constructed plasmid was verified by Polymerase Chain Reaction (PCR) and sequencing (Genscript Biotech Ltd, China).
Cells culture and transfection with sgRNA:
Mouse HSC-T6 was purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd and grew in Dulbeco’s Modified Eagle Media (DMEM) (Gibco, United States of America (USA)). Cell cultures were incubated in a 37° incubator with 5 % Carbon dioxide (CO2) and saturated humidity. Transfection of designed sgRNA plasmid was conducted by using Lipofectamine 3000 (Thermo Fisher, USA).
Effective sgRNA screening:
The plasmid was mixed and then transfected into HSC-T6. When the cells were cultured to grow, the DMEM medium containing puromycin (50 μg/ml) was replaced every 48 h until a positive clonal cell population appeared. After extensive cell expansion, DNA was extracted by genomic DNA extraction kit (Aisijin Biotech Ltd, Shenzhen, China) then PCR was performed. Primers for exon 11 were designed as; ADAM9-Exon11-F: 5’GGTCTGTTGATGCCTGAT3’ and ADAM9- Exon11-R: 5’ATGTAATATGCCCTACCC3’.
PCR was conducted in a 25 μl reaction volume, consisting of 1 μl Oligo-F, 1 μl Oligo-R, 1 μl complementary DNA (cDNA), 12.5 μl 2Es Taq MasterMix, 9.5 μl RNase-free H2O (all from Shanghai Shenggong Biotech Ltd), according to the following conditions: 39 cycles of 94° for 30 sec, 56° for 30 sec and 72° for 30 sec and a 30 min extension step at 72°. After agarose gel electrophoresis, the PCR products were recovered by gel extraction kit (Omega, USA) and the recovered DNA was sent to Shanghai Sheng gong Biotech Ltd for sequencing.
Animals and ALF induction:
Healthy male C57BL/6J mice (SCXK20150001) (about 7-8 w old, 22.5±2.5 g) were purchased from Liaoning Changsheng Biotechnology Co., Ltd (Jinan, Shandong, China). All animals were kept under standard conditions and adapted to the environment for 1 w before the experiment. All experiments were conducted in accordance with the guide for the care and use of laboratory animals.
The 280 mice were randomly divided into the following 4 groups of 70 animals each, as follows; the normal group were fed with control Lieber- DeCarli TP4030C which was purchased from Nantong Trophic Animal Feed High-Tech Co., Ltd (Nantong, China) for 8 w; the saline+alcohol group were fed with Lieber-DeCarli TP4030A and injected with saline; the ADAM9-sgRNA+alcohol group were fed with Lieber-DeCarli TP4030A and injected with effective sgRNA plasmids (50 μg) via tail vein and the JNK inhibitor+alcohol group were fed with Lieber-DeCarli TP4030A and injected with JNK inhibitor SP600125 (24 ml/kg) via tail vein. All mice except for the normal group received intraperitoneal injection of Carbon tetrachloride (CCl4) (2 ml/ kg, twice/week, 5 % in olive oil) from the 5 w of modelling to the 18 w to establish the ALI model. Finally, all mice were sacrificed to collect blood and liver tissues for experiments.
Detection of serum Aspartate Transaminase (AST) and Alanine Transaminase (ALT) activity:
The eyeballs of mice were removed to collect blood and serum was obtained by centrifuging (2500 rpm, 10 min). The activities of ALT and AST in serum were determined by an automatic biochemical analyser. Enzyme activities were expressed as an International Unit per liter (IU/l).
Pathological analysis by Hematoxylin-Eosin (H&E) and picric acid-sirius red staining:
Each group of mice was dissected to obtain liver lobes. The liver lobes fixed in 10 % formaldehyde for 24 h were dehydrated and embedded in paraffin and processed into sections, which were subjected to H&E and picric acid-sirius red staining to evaluate the degree of necrosis and fibrosis of ALF. All sections were examined and pictured under a fluorescence microscope. Areas of necrosis were quantified using Image J v1.8.0 software (National Institutes of Health, Bethesda, MD, USA). According to the degree of liver parenchymal necrosis, the score of hepatocyte necrosis rate was counted (Grade 0: no necrosis; Grade 1: 1 to 2 necrosis; Grade 2: 2 or more necrosis; Grade 3: massive necrosis). According to the degree of collagen fibres in the liver of mice, the Ishak scoring system[16] was used for analysis and the grade of inflammatory activity and the degree of fibrosis were recorded.
Hoechst 33 258 staining to analyse hepatocyte apoptosis:
The sections were routinely dew axed and hydrated, stained with Hoechst staining kit (Beyotime, Shanghai, China) under dark conditions, washed with Phosphate-Buffered Saline (PBS) buffer and sealed with anti-quenching sealing tablets. 10 low power fields were selected under the fluorescence microscope (Olympus, Tokyo, Japan) to observe the staining of the sections and take photos. Calculate the apoptotic rate=(number of apoptotic cells/total number of cells)×100 % and the hepatocyte apoptosis of mice in each group was recorded respectively.
Western blotting in vivo experiment:
Western blotting of p-c-Jun, Cytochrome P450 2E1 (CYP2E1), Tumor Necrosis Factor alpha (TNF-α), Alpha-Smooth-Muscle Actin (α-SMA), Bcl-2-Associated X (BAX), Heat Shock Protein 70 (HSP70), Proliferating Cell Nuclear Antigen (PCNA), ADAM9 and Vascular Endothelial Growth Factor (VEGF).
After the 18 w of alcohol induction, the protein was extracted from the mouse liver and quantified by the Bicinchoninic Acid (BCA) protein assay kit (Solarbio, shanghai, China). After the extracted protein was boiled and denatured, run the electrophoresis according to the size of the factor, transferred the membrane for 2 h and blocked the milk powder for 1 h. The membranes were incubated with primary antibodies targeting ADAM9, PCNA, VEGF, BAX, caspase-3, HSP70, CYP2E1, p-c- Jun, TNF-α and α-SMA (Santa Cruz) at 37° for 1 h. Then the membranes were incubated with secondary antibodies, HRP-conjugated goat or rabbit antimouse Immunoglobulin G (IgG) polyclonal antibody (Zhongshan Jinqiao Biological Ltd, China) at 37° for 1 h and 3,3’-Diaminobenzidine (DAB) (Sigma, USA) was used to show colour. The band density of the target protein was determined by gel pro analyser software 4.0 (Media Cybernetics, Bethesda, MD, USA) and the intensities of the bands were normalized against β-actin.
Western blotting of p-c-Jun, α-SMA, PCNA, ADAM9, Caspase-3 and VEGF in vitro experiment:
HSC-T6 of mice in the logarithmic growth stage were inoculated into 6-well plates and labelled as four groups; normal group, normal+alcohol group, ADAM9-sgRNA+alcohol group (effective sgRNA was stably transfected into cells), JNK inhibitor+alcohol group (JNK inhibitor SP600125 was added to the culture medium at the concentration of 100 ng/ml and the cells were incubated in advance for one night). When the cell density reached 80 %, HSC-T6 in the normal group was cultured normally and the other three groups of cells were treated with 33 μl/ml alcohol for culture. After 24 h, cells from each well were collected to extract protein. Following that, Western blotting of p-c-Jun, α-SMA, PCNA, ADAM9, caspase-3 and VEGF was performed.
Statistical analysis:
All experiments were performed at least three times with triplicate. All data were presented as the mean±Standard Deviation (SD). Statistical comparisons were made using one-way Analysis of Variance (ANOVA) with the Least Significant Difference (LSD)-t test for multiple comparisons. All statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) 16.0 (SPSS Inc., Chicago, Illinois, USA).
Results and Discussion
Design primers on the mouse ADAM9 gene sequence and construct a three-in-one plasmid using CRISPR/Cas9 technology. The plasmid was mixed and then transfected into HSC-T6. Compared with the wild ADAM9 sequence, the sequencing results of ADAM9+sgRNA were quite different by DNAssist software (fig. 1) and it is determined that the target plasmid is successfully constructed.
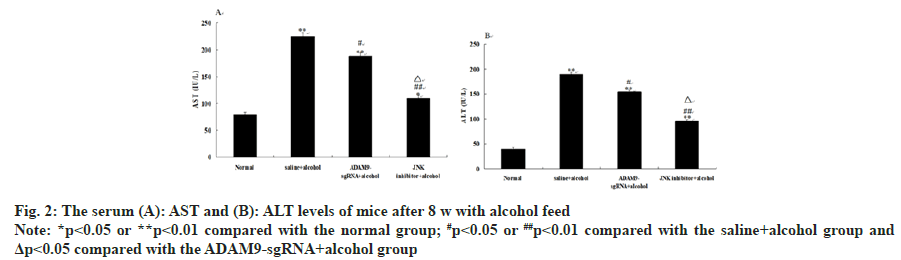
Compared with the saline+alcohol group, the levels of AST and ALT in the ADAM9-sgRNA+alcohol group and the JNK inhibitor+alcohol group significantly decreased (p<0.01 or p<0.05) (fig. 2), while the levels of the JNK inhibitor+alcohol group were significantly lower than those in the ADAM9- sgRNA+alcohol group (p<0.05) (fig. 2). These results suggested that long-term alcohol stimulation could cause liver fibrosis in mice, but inhibiting ADAM9 gene expression and JNK signaling pathway could effectively lower the increase of serum AST and ALT activities.
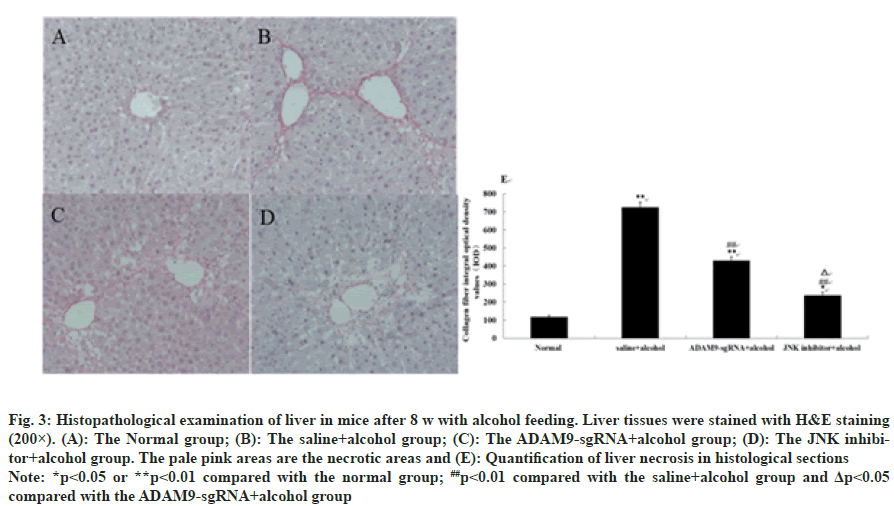
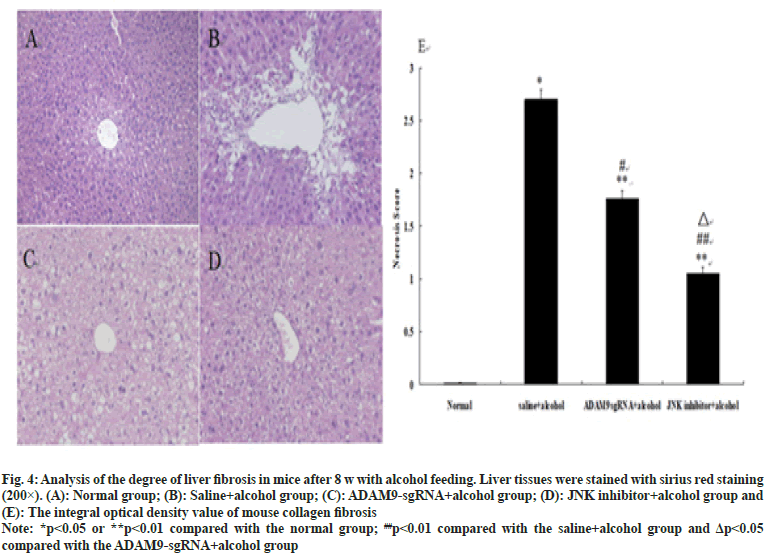
As shown in fig. 3 and fig. 4, different degrees of liver damage was obvious in pathological sections of the mice with alcohol treatment (p<0.01 or p<0.05). With respect to the saline+alcohol group, the liver injury of mice in ADAM9-sgRNA+alcohol group and JNK inhibitor+alcohol group was significantly reduced (p<0.01 or p<0.05) (fig. 3 and fig. 4), while the degree of hepatocyte necrosis and fibrosis of mice in the JNK inhibitor+alcohol group decreased more significantly (p<0.05) (fig. 3 and fig. 4). It was demonstrated that alcohol could lead to severe hepatic injury, but inhibiting ADAM9 gene expression and inhibiting the JNK signaling pathway could evidently alleviate the extent of hepatocyte necrosis and fibrosis.
Fig. 3: Histopathological examination of liver in mice after 8 w with alcohol feeding. Liver tissues were stained with H&E staining
(200×). (A): The Normal group; (B): The saline+alcohol group; (C): The ADAM9-sgRNA+alcohol group; (D): The JNK inhibitor+
alcohol group. The pale pink areas are the necrotic areas and (E): Quantification of liver necrosis in histological sections
Note: *p<0.05 or **p<0.01 compared with the normal group; ##p<0.01 compared with the saline+alcohol group and Δp<0.05
compared with the ADAM9-sgRNA+alcohol group
Fig. 4:Analysis of the degree of liver fibrosis in mice after 8 w with alcohol feeding. Liver tissues were stained with sirius red staining
(200×). (A): Normal group; (B): Saline+alcohol group; (C): ADAM9-gRNA+alcohol group; (D): JNK inhibitor+alcohol group and
(E): The integral optical density value of mouse collagen fibrosis
Note: *p<0.05 or **p<0.01 compared with the normal group; ##p<0.01 compared with the saline+alcohol group and Δp<0.05
compared with the ADAM9-sgRNA+alcohol group
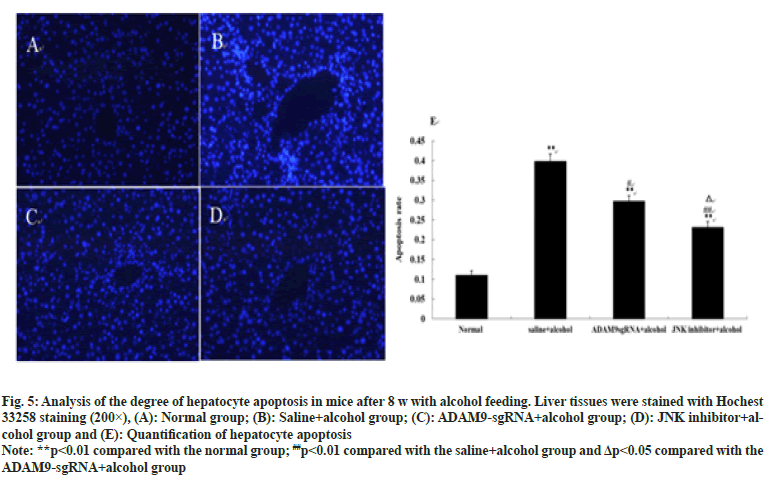
Under the fluorescence microscope, obvious hepatocyte apoptosis was observed in the other three groups of mice except for the normal group (p<0.01), as can be seen in fig. 5. Compared with the saline+alcohol group, the number of hepatocyte apoptosis in ADAM9-sgRNA+alcohol group and JNK inhibitor+alcohol group declined (p<0.05 or p<0.01) (fig. 5), while the number of hepatocyte apoptosis of mice in the JNK inhibitor+alcohol group decreased more significantly (p<0.05) (fig. 5). It was indicated that long-term alcohol stimulation could induce hepatocyte apoptosis, but inhibition of ADAM9 gene expression and JNK signalling pathway could significantly reduce hepatocyte apoptosis.
Fig. 5: Analysis of the degree of hepatocyte apoptosis in mice after 8 w with alcohol feeding. Liver tissues were stained with Hochest
33258 staining (200×), (A): Normal group; (B): Saline+alcohol group; (C): DAM9-sgRNA+alcohol group; (D): JNK inhibitor+alcohol
group and (E): Quantification of hepatocyte apoptosis
Note: **p<0.01 compared with the normal group; ##p<0.01 compared with the saline+alcohol group and Δp<0.05 compared with the
ADAM9-sgRNA+alcohol group
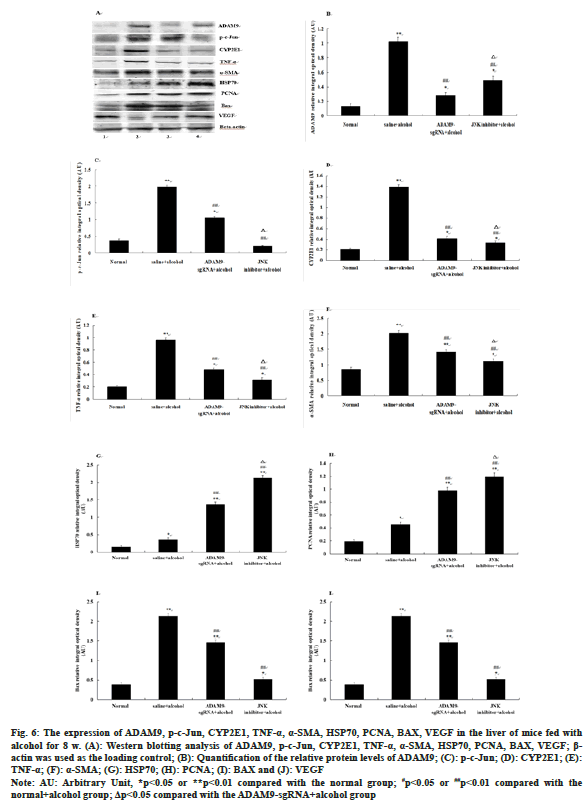
Compared with the saline+alcohol group, the expression of ADAM9, p-c-Jun, CYP2E1, TNF-α, α-SMA and BAX in mice of the ADAM9- sgRNA+alcohol group and the JNK inhibitor+alcohol group all decreased significantly (p<0.05 or p<0.01) (fig. 6A-fig. 6I), while the expression of HSP70, PCNA and VEGF increased significantly (p<0.05 or p<0.01) (fig. 6A-fig. 6J). Among them, ADAM9, HSP70, PCNA and VEGF were significantly expressed higher in mice of the JNK inhibitor+alcohol group than the ADAM9-sgRNA+alcohol group (p<0.05) (fig. 6A, fig. 6B, fig. 6G, fig. 6H, fig. 6J), while the expression of p-c-Jun, CYP2E1, TNF-α and α-SMA was opposite (p<0.05) (fig. 6A, fig. 6C-fig. 6F) and the expression of BAX had no significant difference (fig. 6A, fig. 6I).
Fig. 6: The expression of ADAM9, p-c-Jun, CYP2E1, TNF-α, α-SMA, HSP70, PCNA, BAX, VEGF in the liver of mice fed with
alcohol for 8 w. (A): Western blotting analysis of ADAM9, p-c-Jun, CYP2E1, TNF-α, α-SMA, HSP70, PCNA, BAX, VEGF; β-
actin was used as the loading control; (B): Quantification of the relative protein levels of ADAM9; (C): p-c-Jun; (D): CYP2E1; (E):
TNF-α; (F): α-SMA; (G): HSP70; (H): PCNA; (I): BAX and (J): VEGF
Note: AU: Arbitrary Unit, *p<0.05 or **p<0.01 compared with the normal group; #p<0.05 or ##p<0.01 compared with the
normal+alcohol group; Δp<0.05 compared with the ADAM9-sgRNA+alcohol group
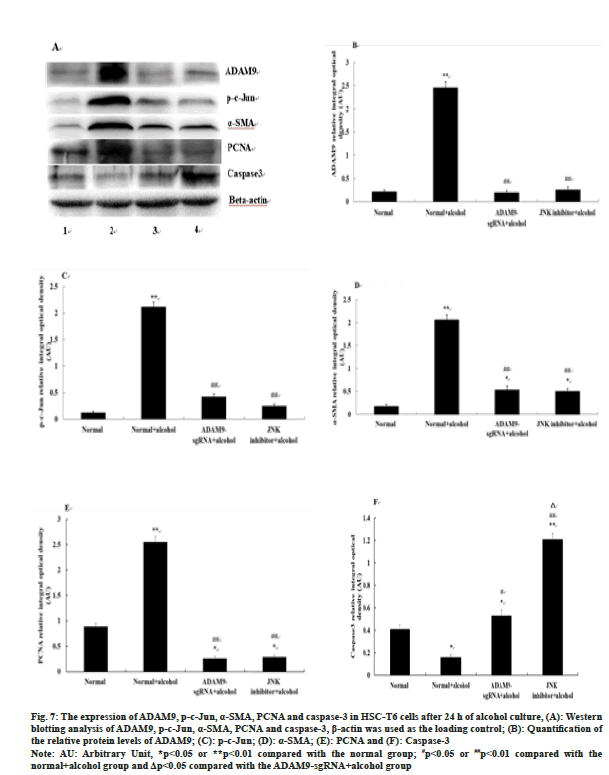
The expression of ADAM9, p-c-Jun, α-SMA and PCNA in the ADAM9-sgRNA+alcohol group and the JNK inhibitor+alcohol group was significantly down-regulated (p<0.05) (fig. 7A-fig. 7E), while caspase-3 expression was up-regulated compared with the normal+alcohol group (p<0.01 or p<0.05) (fig. 7A, fig. 7F). Compared with the ADAM9- sgRNA+alcohol group, caspase-3 expression in the JNK inhibitor+alcohol group increased significantly (p<0.05) (fig. 7A, fig. 7F), while there was no significant difference in the expression of ADAM9, p-c-Jun, α-SMA and PCNA (fig. 7A-fig. 7E).
Fig. 7:The expression of ADAM9, p-c-Jun, α-SMA, PCNA and caspase-3 in HSC-T6 cells after 24 h of alcohol culture, (A): Western
blotting analysis of ADAM9, p-c-Jun, α-SMA, PCNA and caspase-3, β-actin was used as the loading control; (B): Quantification of
the relative protein levels of ADAM9; (C): p-c-Jun; (D): α-SMA; (E): PCNA and (F): Caspase-3
Note: AU: Arbitrary Unit, *p<0.05 or **p<0.01 compared with the normal group; #p<0.05 or ##p<0.01 compared with the
normal+alcohol group and Δp<0.05 compared with the ADAM9-sgRNA+alcohol group
In the current study, we successfully used the plasmid constructed in our laboratory in the early stage and inhibit ADAM9 gene expression of mice[17], which paved the way for the study of the role of ADAM9 in ALF of mice. Through results of ALT and AST, we found the promoting effect of ADAM9 in ALF mice. The pathological analysis further confirmed this finding, which laid a foundation for an in-depth study of the pathogenesis of ALF. It was speculated that ADAM9 could promote ALF via the JNK signal pathway. Further results in vivo and in vitro, it was proved that inhibiting ADAM9 gene expression could alleviate ALF in mice by regulating the JNK signal pathway.
In the progress of ALF in mice, the expression of pro-apoptotic protein BAX increases, which induces hepatocyte apoptosis. Apoptotic hepatocytes form apoptotic bodies that can be engulfed by HSC and cause HSC activation and proliferation. Caspase-3 is called "death protease" and is the executor of apoptosis[18]. After inhibiting the ADAM9 gene expression and injecting JNK inhibitor, the expression of caspase-3 in HSC increased significantly, the pro-apoptotic effect of alcohol on cells reduced and the activation of HSC was inhibited, indicating that ADAM9 could promote hepatocyte apoptosis and the activation of HSC by regulating the JNK signalling pathway and promote the occurrence and development of ALF.
Gehrmann et al.[19] found that the expression level of HSP70 in chronic hepatitis, liver fibrosis, liver cirrhosis and hepatocellular carcinoma showed an upward trend, which had a certain value for the early diagnosis of hepatocellular carcinoma. Alcohol can cause a stress response in mouse liver and increase the expression of HSP70 responsively, stabilizing cell structure and internal environment and protecting cells. After inhibiting ADAM9 gene expression and injecting JNK inhibitor, the expression of HSP70 increased significantly and the protective effect of HSP70 on the liver of mice strengthened, suggesting that ADAM9 could promote ALF via the JNK signal pathway.
The results of PCNA showed that ADAM9 has a dual effect on cell proliferation during ALF in mice. On the one hand, it could inhibit the proliferation of hepatocytes and the repair after liver injury. On the other hand, it could promote the proliferation and activation of HSC and accelerate the growth of the liver. In the process of fibrosis, ADAM9 achieved dual regulation by regulating the JNK signaling pathway.
Studies have confirmed that the JNK plays an important role in the occurrence and development of liver fibrosis[20]. Sustained JNK activation is directly and positively correlated with liver injury and liver metabolic stress dysfunction. Studies have confirmed that a variety of cytokines can activate HSC through the JNK signaling pathway and promote the progress of liver fibrosis. The application of JNK specific blockers can inhibit HSC proliferation and effectively fight liver fibrosis[21]. Under endoplasmic reticulum stress, JNK protein phosphorylates into active p-c-Jun protein. After alcohol activated the JNK signaling pathway, inhibiting ADAM9 gene expression and adding the JNK inhibitor could significantly decrease the expression of p-c-Jun, which effectively reduced the inflammatory response and cell damage in the process of ALF, suggesting ADAM9 could promote HSC activation and hepatic fibrosis by regulating JNK signal pathway.
Acknowledgements:
This work was supported by Central Plains Science and Technology Innovation Leader Project (No. 214200510004), National Natural Science Foundation of China (82170606), Luoyang City Science and Technology Plan Project (2101028A), 2020 Key Scientific Research Project Guidance Plan of Henan Provincial Higher Education (22B3100052), Research and Cultivation Fund of Luoyang polytechnic (SJPY202102) and Luoyang City Science and Technology Plan Project (2101030A). The authors thank all the members in the laboratory when this work was carried out. The experiments comply with the current laws of China.
Conflict of interests:
The authors have no conflict of interests.
References
- Ohashi K, Pimienta M, Seki E. Alcoholic liver disease: A current molecular and clinical perspective. Liver Res 2018;2(4):161-72.
[Crossref] [Google Scholar] [PubMed]
- Thiele M, Detlefsen S, Møller LS, Madsen BS, Hansen JF, Fialla AD, et al. Transient and 2-dimensional shear-wave elastography provide comparable assessment of alcoholic liver fibrosis and cirrhosis. Gastroenterology 2016;150(1):123-33.
[Crossref] [Google Scholar] [PubMed]
- Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 2017;121:27-42.
[Crossref] [Google Scholar] [PubMed]
- Odagiri N, Matsubara T, Sato-Matsubara M, Fujii H, Enomoto M, Kawada N. Anti-fibrotic treatments for chronic liver diseases: The present and the future. Clin Mol Hepatol 2021;27(3):413-24.
[Crossref] [Google Scholar] [PubMed]
- Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, et al. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 2016;7(1):10993.
[Crossref] [Google Scholar] [PubMed]
- Xiong J, Ni J, Chen C, Wang K. miR-148a-3p regulates alcoholic liver fibrosis through targeting ERBB3. Int J Mol Med 2020;46(3):1003-12.
[Crossref] [Google Scholar] [PubMed]
- Stickel F, Moreno C, Hampe J, Morgan MY. The genetics of alcohol dependence and alcohol-related liver disease. J Hepatol 2017;66(1):195-211.
[Crossref] [Google Scholar] [PubMed]
- Breun M, Schwerdtfeger A, Martellotta DD, Kessler AF, Monoranu CM, Matthies C, et al. ADAM9: A novel player in vestibular schwannoma pathogenesis. Oncol Lett 2020;19(3):1856-64.
[Crossref] [Google Scholar] [PubMed]
- Oria VO, Lopatta P, Schilling O. The pleiotropic roles of ADAM9 in the biology of solid tumors. Cell Mol Life Sci 2018;75(13):2291-301.
[Crossref] [Google Scholar] [PubMed]
- Schwettmann L, Wehmeier M, Jokovic D, Aleksandrova K, Brand K, Manns MP, et al. Hepatic expression of A disintegrin and metalloproteinase (ADAM) and ADAMs with thrombospondin motives (ADAM-TS) enzymes in patients with chronic liver diseases. J Hepatol 2008;49(2):243-50.
[Crossref] [Google Scholar] [PubMed]
- Zhang YY, Li SQ, Song Y. Expression analysis of a disintegrin and metalloprotease 9 during acute liver injury induced by alcohol in mice. Chin J Clin Pharmacol 2018;34(18):38-40.
- Li G, Qi W, Li X, Zhao J, Luo M, Chen J. Recent advances in c-Jun N-terminal kinase (JNK) inhibitors. Curr Med Chem 2021;28(3):607-27.
[Crossref] [Google Scholar] [PubMed]
- Hammouda MB, Ford AE, Liu Y, Zhang JY. The JNK signaling pathway in inflammatory skin disorders and cancer. Cells 2020;9(4):857.
[Crossref] [Google Scholar] [PubMed]
- Qia Z. The role of C-Jun N-terminal kinase inhibitor in PDGF-induced autophagy of hepatic stellate cells. J Hebei Med Univ; 2014.
- Qiu AG, Xu SR, Li J, Zhang HM, Gu XH. Protective effect of SP600125 on hepatic ischemia/reperfusion injury in rats. Chin J Tissue Eng Res 2010;14(53):9977-81.
- Zhou BO, Wang G, Gao S, Chen YE, Jin C, Wang Z, et al. Expression of ERO1L in gastric cancer and its association with patient prognosis. Exp Ther Med 2017;14(3):2298-302.
[Crossref] [Google Scholar] [PubMed]
- Zhoa Y. The function of ADAM9 in alcoholic liver injury; 2018.
- Katifelis H, Lyberopoulou A, Mukha I, Vityuk N, Grodzyuk G, Theodoropoulos GE, et al. Ag/Au bimetallic nanoparticles induce apoptosis in human cancer cell lines via P53, CASPASE-3 and BAX/BCL-2 pathways. Artif Cells Nanomed Biotechnol 2018;46(3):S389-98.
[Crossref] [Google Scholar] [PubMed]
- Gehrmann M, Cervello M, Montalto G, Cappello F, Gulino A, Knape C, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Front Immunol 2014;5:307.
[Crossref] [Google Scholar] [PubMed]
- Win S, Than TA, Zhang J, Oo C, Min RW, Kaplowitz N. New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatology 2018;67(5):2013-24.
[Crossref] [Google Scholar] [PubMed]
- Sovolyova N, Healy S, Samali A, Logue SE. Stressed to death–mechanisms of ER stress-induced cell death. Biol Chem 2014;395(1):1-3.
[Crossref] [Google Scholar] [PubMed]