- *Corresponding Author:
- Gayatri Dave
P. D. Patel Institute of Applied Sciences, Charotar University of Science and Technology, Education Campus, Anand, Gujarat 388421, India
E-mail: gayatridave.bt@charusat.ac.in
| Date of Received | 22 June 2021 |
| Date of Revision | 28 October 2022 |
| Date of Acceptance | 12 May 2023 |
| Indian J Pharm Sci 2023;85(3):698-708 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Traditional Indian medicine practice (Ayurveda) emphasized the role of "panchgavya" five products from Bos indicus for human welfare. Ayurveda classics "Sushruta Samhita", "Ashtanga Sangraha" alluded to the therapeutic potential of pristine cow urine as drug or drug ingredients. Compelling evidence exhibits the innumerable medicinal properties of cow urine; accordingly, this elixir can directly treat complex ailments such as leprosy, tuberculosis and fever. Also, the classics narrated many formulations that have utilized cow urine for the preparation of drugs, supplemented to enhance the potency. This practice is more empirical, and only a few pieces of experimental evidence supporting the claim are known. The associated mechanisms are poorly understood and so render its appeal to the limited mass. The study aims to investigate the bio-enhancer-like properties of cow urine toward network pharmacology. For that, 25 medicines having antibacterial, anti-fungal, anti-viral, enzyme inhibitors, and anti-inflammatory actions were selected as a reference. Network analysis for twenty chemotypes found in cow urine was carried out. First, through enrichment analysis, the kyoto encyclopedia of genes and genomes and gene ontology terms were obtained. Second, we performed protein-protein interaction studies to screen more targets. Towards this, the drug-protein and cow urine-protein interaction networks are built separately and processed.
Keywords
Ayurveda, cow urine, network analysis, molecular docking
Ayurveda, an Indian traditional medicine system, has adopted a holistic approach, nucleated towards balancing five life-ruling vital factors. The repertoire includes diet, lifestyle, thought modulation, and herbs' use[1,2]. The combinatorial practice emphasized the progressive restoration of the body through balancing these factors[3]. In Ayurveda, a cow is bestowed; the status of the medical dispensary is a source of beaucoup products considered a boon to humanity. It provides cow dung, urine, milk, curd, and ghee (Indian butter). These are altogether referred to as "panchgavya" in Ayurveda. Cow Urine (CU), usually regarded as a nonessential byproduct in Ayurveda, is used to prepare many herbal formulations. Has medicinal value, a whole or with other active ingredients, can treat ailments, including terminal illness[4]. The benefits of supplemented CU are thoroughly reviewed in Ayurvedic classics Charaka Samhita and Sushruta Samhita (an early journal for surgical practices). In the present era, recent studies conducted at CU Treatment and Research Centre, Indore (India), show the positive association of CU (Gomutra) in the treatment of disorders like blood pressure, artery blockages, and cancer[5]. First records cite its application around 4000 BC; though deep-rooted, its compatibility with the allopathic practice must be established to gain broad acceptance and mass appeal. Despite extensive therapeutic uses, the method is more empirical than experimental and poorly recognized in a region outside the Indian subcontinent. There is a dire need for re-investigation towards established goldstandard research design of biomedical sciences.
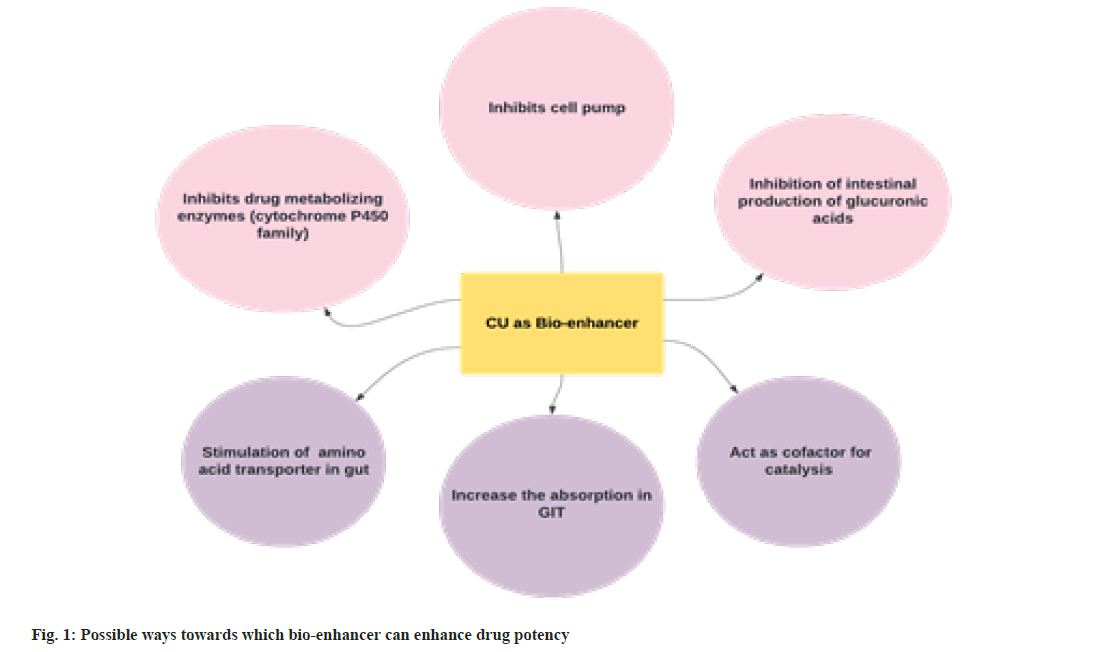
Recently, United States patent[6] (No. 6896907 and 6410059) and a few other laboratories are reportedly claiming bioenhancer properties of CU. Generally, a bio enhancer can be a natural or artificially synthesized substance/s that surges the efficacy of the drug when administered together. It increases the bioavailability of an orally administered pharmaceutical compound through several mechanisms. The bioenhancer often improves the solubility or adsorption of medicine, inhibits the action of drug-metabolizing enzymes[7] and enhances the permeability of the cytoplasmic membrane for drug entry[8]. It's a non-exhaustive list, and a few other speculated ways of activities are presented in fig. 1.
In earlier attempts, the study of Lee et al.[9] noted a steep rise in the anti-inflammatory activity of apigenin when applied with resveratrol. In another study, the potency of Rifampicin greatly improved when supplemented with the naturally occurring organic compound Piperidine; upon addition, the improvement in anti-tuberculosis activity was documented[10].
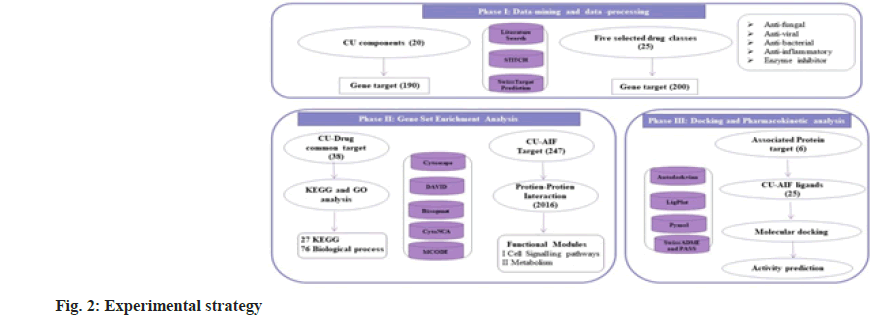
Ayurveda treatment regime advocates the application of whole CU or CU distillate. CU is a rich source of bioactive components known to influence multiple biological pathways toward interacting with diverse biological targets. Viewing multi-target impact Network pharmacology (NP) is the best method to investigate as it deals with numerous networks and components systematically[11]. Bioenhancer, a collective term given to the compounds that enhance the potency/ efficiency or solubility of the drug when administered. To elucidate the role of CU as a bio-enhancer using NP, we first screened 20 chemical components from CU. Unique (limited to certain species) and conditions specific chemicals constituents excluded. Next, we have chosen drugs from five distinct functional categories and such are anti bacterial, anti-fungal, anti-viral, enzyme inhibitors and Anti-Inflammatory (AIF). First, a network for 38 common targets of drugs (25) and CU components (20) is visualized. Overlapping targets annotated through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Next, towards another strategy, we built a CU-AIF core target-Protein Protein Interaction (PPI) network and functional modules obtained. Finally, we curtailed the six key targets showing the role of CU in drug metabolism and cell signaling pathways. Molecular docking and pharmacokinetic analysis studies elucidate the inhibition of the drug-metabolizing enzyme by CU components. Fig. 2 represents the overall experimental approach.
Materials and Methods
The addition of CU to the drug could augment the potency of the drug, to comprehend the impact, the study was conducted by taking two different approaches. First, the overlapping gene targets are fetched from CU and medicines (from all five classes). In the second attempt, We selected only AIF drugs and conducted the PPI and topological analysis to screen other related targets. The conventional data processing and preparation methods are typical for both approaches and are narrated in subsequent sections.
Chemotype processing and target prediction:
A few reports citing the chemical nature of CU have been published so far; patents, reported literature and ayurvedic records through keyword CU names of chemical compounds were obtained. Next, sorting the chemicals into specific chemical classes, their canonical SMILES were collected from PubChem (https://pubchem.ncbi.nlm.nih. gov/)[12] and Zinc (http://zinc15.docking.org/)[13] subsequently converted to SDF formats (Table 1). Through considering the mechanism of action and commercial applications, drugs from five categories, anti bacterial, anti fungal, antiviral, AIF and enzyme inhibitors were chosen to investigate the bioenhancer properties of CU.
| Benzoids | Homogeneous non-metal compound |
|---|---|
| Benzoic acid | Chloride |
| p-Cresol | Copper |
| Phenol | Phosphate |
| Gallic acid | Sulfate |
| Salicylic acid | Nitrite |
| Organic oxygen compound | Phosphorus Pentoxide |
| Lactose | Organo heterocyclic compound |
| Organic acid and derivatives | Uric acid |
| Creatinine | Allantoin |
| Lipid and lipid-like molecule | Nicotine |
| Thymol | Phenylpropenoids and polyketides |
| Ferulic acid | |
| Caffeic acid |
Table 1: Chemotypes Commonly Present in Cow Urine; Class-Wise Summary
Putative gene targets for CU components and medicines were retrieved from STITCH (http:// stitch.embl.de/)[14] and Swiss Target Prediction (http://www.swisstargetprediction.ch/)[15]. Hits with a confidence score of 0.9 to 1 were considered and converted to their corresponding UniProtKB ID (https://www.uniprot.org/) for gene annotation. The list was re-organized and manually curated using published experimental records. Specifically, the gene targets involved in membrane transports, such as ABC transporters and cell proton pumps, were counted.
PPI networks and functional clusters:
Towards strategy II the network for CU and AIF, PPI was visualized. A PPI network built through Bisogenet, a plugin of Cytoscape. Bisogenet is three tire application for investigating the bimolecular relationships. It combines the information from six central PPI databases viz., Molecular Interaction Database (MINT), IntAct Molecular Interaction Database (IntAct), Database of Interacting Proteins (DIP), Human Protein Reference Database (HPRD), Biomolecular Interaction Network Database (BIND), and Biological General Repository for Interaction Datasets (BioGRID)[16]. Input entities' protein identifiers for Homo sapiens were selected in Bisogenet, gene identifiers were uploaded, and the distance from the input node was adjusted to 1. In the output column, only protein identifiers have opted. Next, through an intersection, merged PPI networks of CU-AIF targets were visualized. Subsequently, we used CytoNCA, a plugin of Cytoscape, to identify the essential proteins from the merged network. To determine the node present at a critical location, six centrality measures were selected in CytoNCA[17]. An analysis for significant centrality values such as Degree Centrality (DC), Closeness Centrality (CC), Network Centrality (NC), Betweenness Centrality (BC), Local Average Connectivity-based method (LAC), and Eigenvector Centrality (EC) was conducted.
Minimal Common Oncology Data Elements (MCODE)[18,19], a cluster analysis algorithm in Cytoscape, detects the densely-connected area in PPI networks. MCODE effectively builds a molecular interaction network based on connectivity.
Functional annotation of hits of strategy I and strategy II:
In a first attempt, 38 overlapping hits were uploaded to Database for Annotation, Visualization, and Integrated Discovery (DAVID)[20] to find out the enriched terms through KEGG[21] and GO[22] databases. Hits with a p-value of 0.005 were selected for achieving the list of significant targets. We used the ImageGP gene enrichment tool to visualize enriched GO and KEGG terms. Second, the hits of PPI clusters were analyzed and information on associated biological pathways was obtained. KEGG and GO terms from the strategy I and II were screened for repetitive entries and repeated names were pooled for subsequent analysis.
Structure-based docking and activity predictions:
The overlapping GO and KEGG terms from both the strategies were screened and further investigated for their drug binding affinities. Frail interaction of a protein-ligand was weed out through performing molecular docking in the Autodock vina ver 4.1[23]. The X-ray crystal structures for selected targets; CYP1A1(PDB ID:6DWM), CYP1A2(PDB ID: 2HI4), UGT1A3(PDB ID: 1V4T), ABCB1(PDB ID: 6GDI), CYP3A4(PDB ID: 4I3Q), CYP2D6(PDB ID: 2F9Q), COX-1(PDB ID:6Y3C) and COX- 2 (PDB ID:5KIR) were obtained from research collaboratory for structural bioinformatics protein data bank (www.rcsb.org). Ligand-protein chemical interactions scored through visualization and interaction analysis in PyMol 1.8[24] and LigPlot v 4.5.3[25]. Using Prediction of Activity Spectra for Substances (PASS) software, we predicted the pharmacokinetic activities for CU and drugs. PASS allows us to predict the mechanism of action a ligand have on protein targets; results were re-confirmed with SwissADME (http://www. swissadme.ch/) pharmacokinetic analysis [26].
Results and Discussion
CU can act as bioenhancer. The present study investigates the hypothesis through NP tools. Towards it, through an extensive literature review, two ligand datasets were generated. One represents CU compounds containing 20 distinct chemotypes (dataset-1), and the other has the list of lead compounds of five commercially available drugs (dataset-2). Drugs having an analogous mechanism of action were scrutinized manually through published reports. From a class anti-viral, medicines Benazepril and Captopril inhibit the conversion of angiotensin I to angiotensin II. They share a similar protein target and are mainly taken to enrich the maximum genes from biological pathways associated with the various medicines of the same class. The selected categories of drugs are known to act on numerous macromolecules from bacteria, fungi, viruses and Homo sapiens. Our study limits human macromolecular targets; this reduces the noise in the analysis at the end.
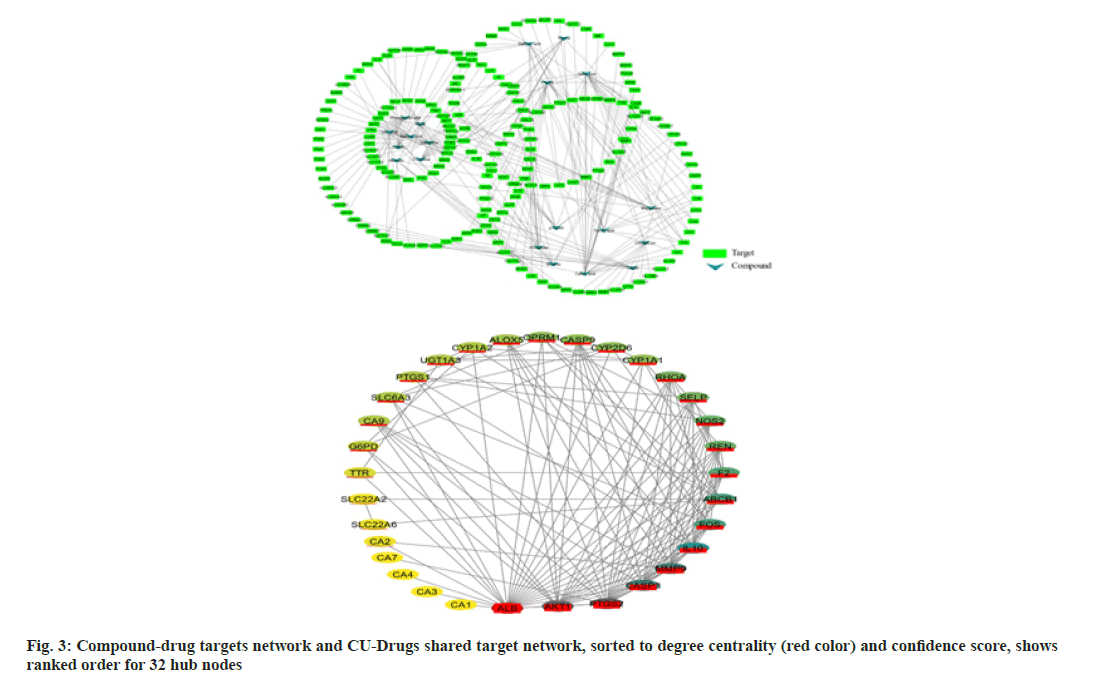
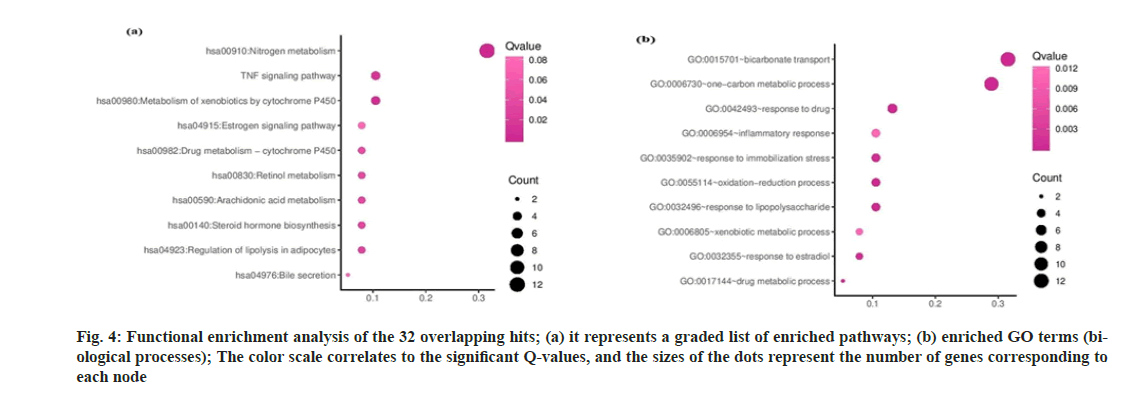
A ligand-target network constructed through Cytoscape (https://cytoscape.org/;version 3.8.0)[27] shows a reasonably extensive network with 208 nodes and 234 edges (fig. 3a). Post abandoning non-overlapping hits,38 commonhits were curtained and presented in fig. 3b, describing 32 hub nodes and 123 edges. The hub nodes having edges five or above were manually scrutinized and selected for further analysis. The thirty-eight annotated targets were associated with 76 biological processes, 25 molecular functions, 15 cellular components and 27 KEGG pathways. The results indicate that the CU-associated targets are mainly located on the plasma membrane and involved in the metabolism and transport of small molecules, evident from gene enrichment plots. We only considered the hits with Q-value≤0.05 (fig. 4a and fig. 4b). CU components are generally enriched in the following KEGG pathways:hsa00140:Steroid hormone biosynthesis, hsa00980:Metabolism of xenobiotics by cytochrome P450 hsa00982:cytochrome P450drug-metabolism pathways. The following genes, UGT1A3, CYP1A1, CYP2D6, and CYP1A2, are acting for both CU and drugs in the pathways mentioned above.
Fig. 4: Functional enrichment analysis of the 32 overlapping hits; (a) it represents a graded list of enriched pathways; (b) enriched GO terms (biological processes); The color scale correlates to the significant Q-values, and the sizes of the dots represent the number of genes corresponding to each node
Cellular processes are linked and accomplished by the coordinated action of functional modules.
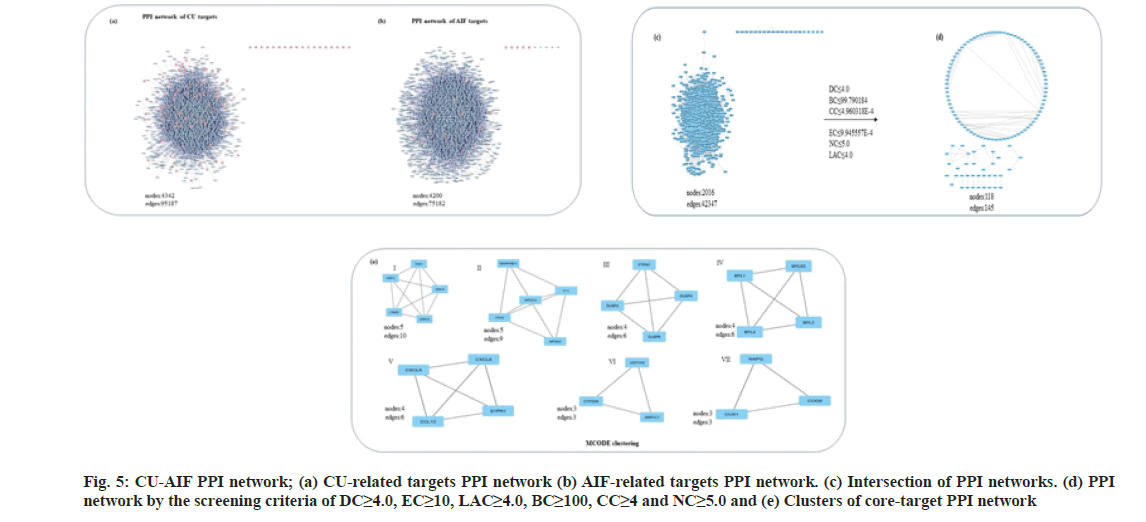
It encompasses the group of proteins involved in catering to similar functions. Towards strategy- II, the PPI networks of CU and AIF-associated proteins developed through Bisogenet (fig. 5a and fig. 5b). The same nodes and edges from the two separate PPI networks were selected to obtain intersection, a merged network containing 2016 hub nodes and 42347 edges (fig. 5c), which is still complex to interpret. Post-screening through six-criterion-based topological analysis in CytoNCA, 200 significant nodes were obtained (fig. 5d). Next, the MCODE clustering analysis gave seven significant clusters (fig. 5e). Proteins from the modules I-V and VII are enriched in cell signaling.
In contrast, the proteins from module VI are associated with the process of drug metabolisms. Overall, the results suggest that the majority hits enriched to two cellular processes; cell signaling and drug metabolism (Table 2). Similar observations strike out from the analysis conducted to the strategy I; Hence a merged list of targets from both approaches curtailed contains CYP2D6, UGT1A3, CYP3A4, ABCB1, CYP1A1, and CYP1A2.
| Cluster classification | Gene targets | |
|---|---|---|
| p-value | Associated protein targets | |
| I Signaling pathways | ||
| hsa04020: Calcium signaling pathway | 2.00×10-3 | GNA15, LTB4R2, ADRA1A |
| hsa04080: Neuroactive ligand-receptor interaction | 4.70×10-3 | P2RY2, LTB4R2, ADRA1A |
| hsa04062: Chemokine signaling pathway | 2.10×10-3 | CCL13, CXCL5, CXCL6 |
| hsa04060: Cytokine-cytokine receptor interaction | 3.60×10-3 | CCL13, CXCL5, CXCL6 |
| hsa04010: MAPK signaling pathway | 4.90×10-5 | PTPN7, DUSP4, DUSP2, DUSP6 |
| hsa00140: Steroid hormone biosynthesis | 1.70×10-2 | UGT1A3, AKR1C1 |
| II Metabolism | ||
| hsa00980: Metabolism of xenobiotics by cytochrome P450 | 1.10×10-4 | UGT1A3, CYP2D6, AKR1C1 |
| hsa00982: Drug metabolism - cytochrome P450 | 2.00×10-3 | UGT1A3, CYP2D6 |
Table 2: List of Significant Pathways and Genes Obtained Through Clustering and Enrichment Analysis
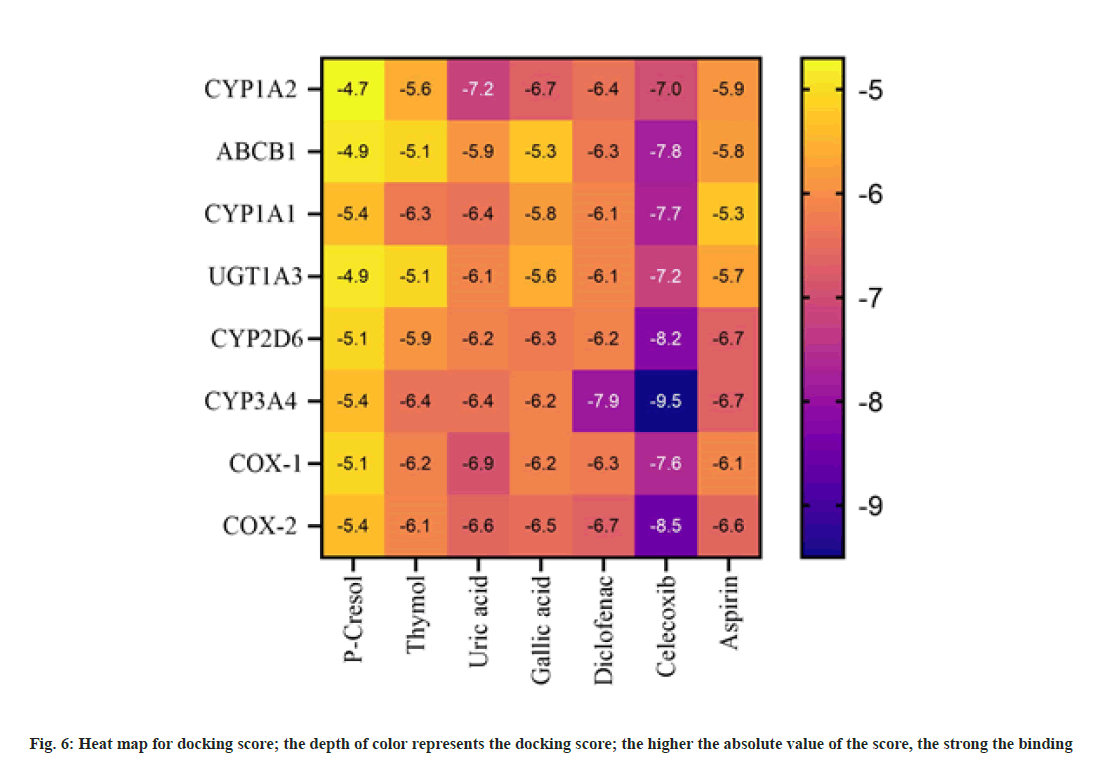
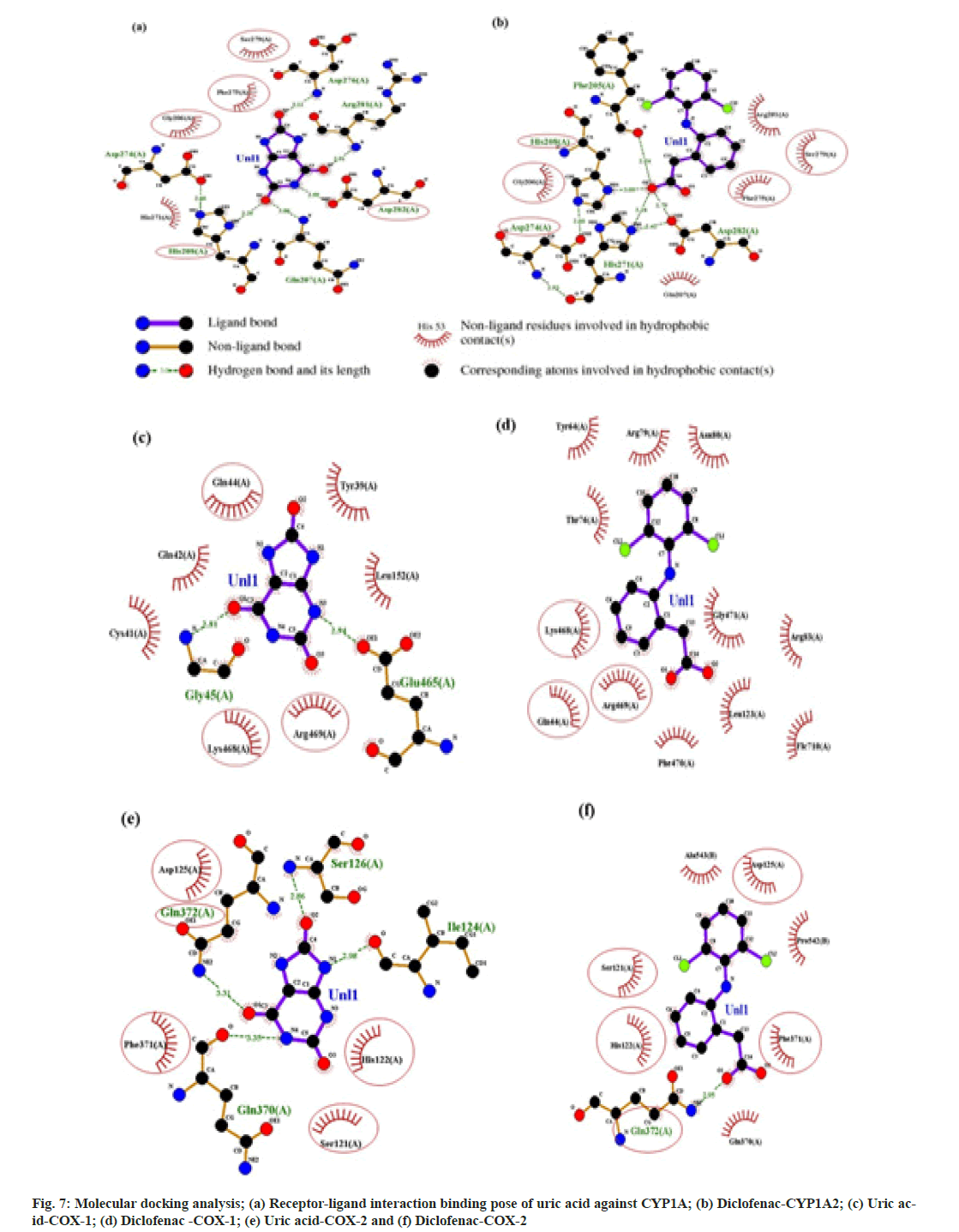
From 4342 CU targets, performing ligandtarget docking weak interaction was weed out. Anti-inflammatory drugs relieve pain and inflammation by inhibiting the cellular cyclooxygenase (COX1 and COX2) activity; here, we selected Celecoxib, diclofenac, and Aspirin as reference drugs. Post docking, the ligands were classified according to the binding affinities shown in fig 6. The lowest binding free energy and the number of H-bonds accounted for the analysis. The binding energy of Uric acid (-7.2 kcal/mol) is compatible with the standard drug diclofenac (-6.4 kcal/mol). For target CYP1A2, diclofenac and Uric acid use the same binding pocket evident from shared interaction with following amino acid residues; Ser279, Phe275, Gly206, His208, and Asp282 (fig. 7A and fig. 7B). This result also suggests that uric acid is a better candidate for the tar get CYP1A2 than diclofenac. For the prime drug target COX- 1 the binding affinity of uric acid (-6.6 kcal/mol) is identical to diclofenac (-6.7 kcal/mol), and again, a similar active site pocket where Gln144, Lys468, and Arg469 are interacting with ligands and showing similar H-bond profiles (fig. 7c and fig. 7d). For another drug target, COX-2, the binding affinities of uric acid (-6.9 kcal/mol) are more significant than diclofenac (-6.3 kcal/ mol), having the following common residues at binding pockets Gln372, Phe371, Ser121, His122 (fig. 7e and fig. 7f).
Pharmacokinetic analysis of screened targets presents a promising role of CU in drug formulations. It suggests that the CU component, gallic acid downregulates the expression of CYP3A4 and thereby delays the drug metabolism[28,29] that further increasing the drug retention time in the body, eventually may result in increases in gastrointestinal absorption of a drug. Another Cu component, p-cresol, and Thymol inhibits the activity of CYP1A2 and CYP2D6[30,31] and exerts similar activity to gallic acid. P-cresol in CU inhibits the membrane p-gp efflux by facilitating the optimum drug delivery in the cell interior[32,33]. Benzoic acid inhibits the activity of CYP1A1, and Uric acid regulates the activity of UGT1A3 in drug metabolism [34,35].
In a grim scenario of the evolution of drug resistance and drug insensitivity, there is a need for quick and affordable solutions[36,37]. The microbes acquire resistance through metabolizing the drug that poses a severe threat to existing therapeutics[38]. Discovering a novel drug entirely to combat this unforeseen change in the behavior of bacteria or cells is an economically challenging and lengthy process[39]. The science of drug designing is perturbed by the rapid emergence of drug resistance towards established antibiotics. The discipline demands an urgent need for repositioning or reformulating the current drugs. Researcher across the globe is testing various strategies such as the inclusion of metabolic inhibitors[40] to singlegene deletion[41,42]. Adding bioenhancer may be one more alternative that may affect the drug metabolism[43-45]. During the literature analysis for Ayurveda practice, we observed the practice of inclusion of CU distillate in many Ayurveda formulation, has numerous application and exert its impact through modulating the fundamental processes[46].
Drug effectiveness depends on several biological processes, including membrane transport/ binding to blood plasma proteins/vasodilation of human intestinal tight junctions/endocytosis[47]. These processes mainly exalt drug absorption and distribution in the human body. The pharmacokinetics of any drug is related to many such biological processes. The results of studies conducted for the strategy I and II show that drug metabolism, one-carbon metabolism, response to the drug, cellular transports, and cell signaling pathways are shared among all the selected drugs associated with CU components. Though it is in silico studies, it firmly establishes the interaction of CU with key gene targets that are often essential for drug activity. Hence, CU could be introduced as a bio enhancer. Laboratory validation studies are imperative to prove this claim further.
The elaborated results of docking and pharmacokinetic analysis display the strong association of CU with gene targets CYP2D6, UGT1A3, CYP3A4, ABCB1, CYP1A1, and CYP1A2. Typically, their gene products metabolize the drugs and facilitate their excretion for the human body. Cytochromes P450 (CYPs) are a superfamily of monooxygenase enzymes that oxidize steroids, xenobiotics, and drugs and play a role in its clearance[48-50]. Activation or inhibition of CYP1A1, CYP2D6 CYP3A4, and CYP1A2 isoforms usually activate or prevent the detoxification of numerous xenobiotics. It is one mechanism that regulates the bioavailability of drugs. The CU components interact with these isoforms, and few are involved in its suppression, which reduces the rate of drug metabolism. The reduced rate of drug metabolism increases drug absorption and sometimes drug toxicity .
Uridine diphosphate glucuronosyltransferase 1A3 (UGT1A3) belongs to the uridine diphosphate glucuronosyltransferase superfamily. In a mammal liver, the enzymes predominantly detoxify the endobiotic and xenobiotics and transform them into more polar and water-soluble glucuronides[51,52]. These altered metabolites are biologically inactive and easily extracted from the body. Drugs or herbs that inhibit some members of this family may lead to the accumulation of such molecules in the body, and even sometimes, clinical toxicity is observed. Though the inclusion of bio-enhancer in a drug formulation is a wise choice, it needs optimization to a certain extent; however, the in silico studies exhibit the multilevel impact of CU on drug tar gets.
The use of CU in folklore date back to 4000 BC; however, limited knowledge of pharmacologically active compounds renders its application. It's a prima facie report that supports the ages-old Ayurveda practice that uses CU to prepare many herbal formulations. For target exploration and bioactive mining, NP currently relies on various databases. Despite being curated, databases may show discrepancies due to multiple sources of information, theoretical data, and experimental results. Though NP can accurately predict the genome-wide association of a selected compound, further laboratory investigations are required to elucidate the activity of CU's component towards the drugmetabolizing enzymes.
Furthermore, cows' diet affects the chemical composition of urine, so the chemical constituents must be validated before use. In some cases, the issue can be resolved by fixing the diet of bovines whose urine will be used for medicinal purposes. In the future, detailed pharmacokinetic and pharmacodynamic studies could be designed based on observations of the present manuscript.
Author Contributions:
I want to thank Charotar University of Science and Technology for providing infrastructure.
Conflict of interests:
The authors declared no conflict of interests.
References
- Govindaraj P, Nizamuddin S, Sharath A, Jyothi V, Rotti H, Raval R, et al. Genome-wide analysis correlates Ayurveda Prakriti. Sci Rep 2015;5(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Choudhary N, Singh V. Insights about multi-targeting and synergistic neuromodulators in Ayurvedic herbs against epilepsy: Integrated computational studies on drug-target and protein-protein interaction networks. Sci Rep 2019;9(1):10565.
- Peterson CT, Lucas J, John-Williams LS, Thompson JW, Moseley MA, Patel S, et al. Identification of altered metabolomic profiles following a panchakarma-based ayurvedic intervention in healthy subjects: The self-directed biological transformation initiative (SBTI). Sci Rep 2016;6(1):32609.
- Dudhatra GB, Mody SK, Awale MM, Patel HB, Modi CM, Kumar A, et al. A comprehensive review on pharmacotherapeutics of herbal bioenhancers. Scientific World J 2012;2012:637953.
[Crossref] [Google Scholar] [PubMed]
- Adhikari N, Joshi DR. Cow urine: The Total research on cow urine from the very beginning to till date.
- Randhawa GK, Sharma R. Chemotherapeutic potential of cow urine: A review. J Intercult Ethnopharmacol 2015;4(2):180-6.
[Crossref] [Google Scholar] [PubMed]
- Javed S, Ahsan W, Kohli K. The concept of bioenhancers in bioavailability enhancement of drugs–a patent review. J Sci Lett 2016;1:143-65.
- Peterson B, Weyers M, Steenekamp JH, Steyn JD, Gouws C, Hamman JH. Drug bioavailability enhancing agents of natural origin (bioenhancers) that modulate drug membrane permeation and pre-systemic metabolism. Pharmaceutics 2019;11(1):33.
[Crossref] [Google Scholar] [PubMed]
- Lee JA, Ha SK, Cho E, Choi I. Resveratrol as a bioenhancer to improve anti-inflammatory activities of apigenin. Nutrients 2015;7(11):9650-61.
[Crossref] [Google Scholar] [PubMed]
- Nageswari AD, Rajanandh MG, Uday MK, Nasreen RJ, Pujitha RR, Prathiksha G. Effect of rifampin with bio-enhancer in the treatment of newly diagnosed sputum positive pulmonary tuberculosis patients: A double-center study. J Clin Tuberc Other Mycobact Dis 2018;12:73-7.
[Crossref] [Google Scholar] [PubMed]
- Zhang RZ, Yu SJ, Bai H, Ning K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep 2017;7(1):2821.
- Kim S, Thiessen PA, Cheng T, Yu B, Shoemaker BA, Wang J, et al. Literature information in PubChem: Associations between PubChem records and scientific articles. J Cheminform 2016;8:1-5.
[Crossref] [Google Scholar] [PubMed]
- Sterling T, Irwin JJ. ZINC 15–ligand discovery for everyone. J Chem Inf Model 2015;55(11):2324-37.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res 2016;44(D1):D380-4.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(W1):W357-64.
[Crossref] [Google Scholar] [PubMed]
- Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: A new tool for gene network building, visualization and analysis. BMC Bioinformatics 2010;11:1-9.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
[Crossref] [Google Scholar] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4(1):1-27.
[Crossref] [Google Scholar] [PubMed]
- Xiong Y, Yang Y, Xiong W, Yao Y, Wu H, Zhang M. Network pharmacology‐based research on the active component and mechanism of the antihepatoma effect of Rubia cordifolia L. J Cell Biochem 2019;120(8):12461-72.
[Crossref] [Google Scholar] [PubMed]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007;35(suppl_2):W169-75.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25(1):25-9.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Yuan S, Chan HS, Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip Rev Comput Mol Sci 2017;7(2):e1298.
- Laskowski RA, Swindells MB. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51(10):2778-86.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Pu QH, Shi L, Yu C. Time-dependent inhibition of CYP3A4 by gallic acid in human liver microsomes and recombinant systems. Xenobiotica 2015;45(3):213-7.
[Crossref] [Google Scholar] [PubMed]
- Kollipara S, Gandhi RK. Pharmacokinetic aspects and in vitro–in vivo correlation potential for lipid-based formulations. Acta Pharm Sin B 2014;4(5):333-49.
[Crossref] [Google Scholar] [PubMed]
- Ekins S, Ring BJ, Grace J, McRobie-Belle DJ, Wrighton SA. Present and future in vitro approaches for drug metabolism. J Pharmacol Toxicol Methods 2000;44(1):313-24.
[Crossref] [Google Scholar] [PubMed]
- Chen XW, Serag ES, Sneed KB, Zhou SF. Herbal bioactivation, molecular targets and the toxicity relevance. Chem Biol Interact 2011;192(3):161-76.
[Crossref] [Google Scholar] [PubMed]
- Lim HY, Ho QS, Wong KP. Interplay of metabolizing enzymes and transporter of xenobiotics. Xenobiotica 2016;46(1):25-33.
[Crossref] [Google Scholar] [PubMed]
- Meesters RJ, Duisken M, Hollender J. Cytochrome P450-catalysed arene-epoxidation of the bioactive tea tree oil ingredient p-cymene: Indication for the formation of a reactive allergenic intermediate? Xenobiotica 2009;39(9):663-71.
[Crossref] [Google Scholar] [PubMed]
- Iwai M, Maruo Y, Ito M, Yamamoto K, Sato H, Takeuchi Y. Six novel UDP-glucuronosyltransferase (UGT1A3) polymorphisms with varying activity. J Hum Genet 2004;49(3):123-8.
[Crossref] [Google Scholar] [PubMed]
- Lu J, Shang X, Zhong W, Xu Y, Shi R, Wang X. New insights of CYP1A in endogenous metabolism: a focus on single nucleotide polymorphisms and diseases. Acta Pharm Sin B 2020;10(1):91-104.
[Crossref] [Google Scholar] [PubMed]
- Cheng X, Lv X, Qu H, Li D, Hu M, Guo W, et al. Comparison of the inhibition potentials of icotinib and erlotinib against human UDP-glucuronosyltransferase 1A1. Acta Pharm Sin B 2017;7(6):657-64.
[Crossref] [Google Scholar] [PubMed]
- Kalow W. Ethnic differences in drug metabolism. Clin Pharmacokinet 1982;7(5):373-400.
[Crossref] [Google Scholar] [PubMed]
- Qin X, Wang X. Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharm Sin B 2019;9(6):1087-98.
[Crossref] [Google Scholar] [PubMed]
- Al-Baqsami ZF, Ahmad S, Khan Z. Antifungal drug susceptibility, molecular basis of resistance to echinocandins and molecular epidemiology of fluconazole resistance among clinical Candida glabrata isolates in Kuwait. Sci Rep 2020;10(1):6238.
[Crossref] [Google Scholar] [PubMed]
- Twarog NR, Connelly M, Shelat AA. A critical evaluation of methods to interpret drug combinations. Sci Rep 2020;10(1):5144.
- Elkashif A, Seleem MN. Investigation of auranofin and gold-containing analogues antibacterial activity against multidrug-resistant Neisseria gonorrhoeae. Sci Rep 2020;10(1):1-9.
- Lv C, Huang L. Xenobiotic receptors in mediating the effect of sepsis on drug metabolism. Acta Pharm Sin B 2020;10(1):33-41.
[Crossref] [Google Scholar] [PubMed]
- Tyzack JD, Kirchmair J. Computational methods and tools to predict cytochrome P450 metabolism for drug discovery. Chem Biol Drug Des 2019;93(4):377-86.
[Crossref] [Google Scholar] [PubMed]
- Bora-Singhal N, Mohankumar D, Saha B, Colin CM, Lee JY, Martin MW, et al. Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci Rep 2020;10(1):4722.
[Crossref] [Google Scholar] [PubMed]
- Tatiraju DV, Bagade VB, Karambelkar PJ, Jadhav VM, Kadam V. Natural bioenhancers: An overview. J Pharmacogn Phytochem 2013;2(3):55-60.
- Randhawa GK, Kullar JS. Bioenhancers from mother nature and their applicability in modern medicine. Int J Appl Basic Med Res 2011;1(1):5-10.
[Crossref] [Google Scholar] [PubMed]
- Randhawa G. Cow urine distillate as bioenhancer. J Ayurveda Integr Med 2010;1(4):240.
[Crossref] [Google Scholar] [PubMed]
- Mohanty I, Senapati MR, Jena D, Palai S. Diversified uses of cow urine. Int J Pharm Pharm Sci 2014;6(3):20.
- Seden K, Dickinson L, Khoo S, Back D. Grapefruit-drug interactions. Drugs 2010;70:2373-407.
- Pratanwanich N, Lió P. Pathway-based Bayesian inference of drug–disease interactions. Mol Bio Systems 2014;10(6):1538-48.
- Jiang L, Liang SC, Wang C, Ge GB, Huo XK, Qi XY, et al. Identifying and applying a highly selective probe to simultaneously determine the O-glucuronidation activity of human UGT1A3 and UGT1A4. Sci Rep 2015;5(1):9627.
[Crossref] [Google Scholar] [PubMed]
- Lv X, Xia Y, Finel M, Wu J, Ge G, Yang L. Recent progress and challenges in screening and characterization of UGT1A1 inhibitors. Acta Pharm Sin B 2019;9(2):258-78.
[Crossref] [Google Scholar] [PubMed]