- *Corresponding Author:
- H. L. Raghavendra
Department of Pharmacology
HSK College of Pharmacy, BVVS Campus
Bagalkot-587101, India
E-mail: hallur2018@gmail.com
| Date of Received | 14 September 2020 |
| Date of Revised | 02 February 2021 |
| Date of Acceptance | 10 March 2021 |
| Indian J Pharm Sci 2021;83(2):262-268 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Recent epidemiological studies suggest that postprandial hyperglycemia is an independent risk factor for cardiovascular disease. A human’s high carbohydrate diet majorly consists of a high carbohydrate diet and α-glucosidase is a glucosidase located in the brush border of the small intestine is involved in glycosidic cleavage of starch at α-glycosidic bonds. α-glucosidase inhibitors are a unique class of anti-diabetic drugs particularly useful in individuals with a high carbohydrate diet. α-glucosidase inhibitors works by competitively inhibiting the enzyme α-glucosidase at the brush border of the small intestines, thus delaying the digestion of complex carbohydrate and intestinal absorption of glucose. Hence, in the present study, the α-glucosidase inhibition activity of butyl isobutyl phthalate isolated from chloroform fraction of Rubus steudneri leaves investigated and its possible mechanism of action ascertained in silico. The compound was found to exhibit concentration-dependent inhibition of α-glucosidase with half-maximal inhibitory concentration value of 10.68 g/ml. The results of in vitro α-glucosidase inhibition assay for butyl isobutyl phthalate were promising and substantiated by molecular docking and molecular dynamics studies.
Keywords
Rubus steudneri, diabetes mellitus, molecular docking and dynamics, cresset flare.
The preliminary step in the significant declination of glucose homeostasis is the loss of postprandial glycemic control, which can be progressed to morning hyperglycemia and sustained nighttime hyperglycemia eventually [1]. Impaired glucose tolerance is considered as a pre-diabetic stage, and it may occur years before elevated fasting plasma glucose (FPG) levels are observed [2]. Postprandial plasma glucose (PPG) is a measure to understand the glucose tolerance in a system after the consumption of high carbohydrate meal [3]. There is a growing evidence suggesting that postprandial hyperglycemia may be an independent risk factor for cardiovascular disease, stroke, retinopathy, renal failure and neurologic complications in both diabetic and non- diabetic individuals [1]. Postprandial hyperglycemia is therefore one of the very important pathophysiological states contributing to vascular failure. Accordingly, controlling postprandial hyperglycemia should be the focus of future clinical investigation as a potential target for preventing vascular failure [4].
One such a potential target is α-glucosidase, which is exoenzyme acting in a manner similar to that of glucoamylase on di and oligo saccharides. α-glucosidase inhibitors (AGIs) are particularly useful in individuals whose meals consist of high carbohydrate content. It works by competitively inhibiting the enzyme α-glucosidase at the brush border of the small intestines, thus delaying the digestion of complex carbohydrate and intestinal absorption of glucose [5]. It targets mainly the postprandial hyperglycemia, and in clinical studies, it lowers the HbA1c by 0.5-1.0 %. Gastrointestinal side effects are common andlikely due to colonic starch fermentation. Acarbose is the most widely prescribed α-glucosidase inhibitor; others include Miglitol and Voglibose [6].
There is serious complication of using these AGIs, which include diarrhea, mucus discharge, rectal bleeding and constipation [7]. Therefore, it is necessary to search for alternatives that can display α-glucosidase inhibitory activity but without side reactions. In recent years, projects undertaken to discover potent non- sugar based α-glucosidase inhibitors from natural sources have received tremendous attention because of the highly abundant compounds in nature and their promising biological activities [8].
Rubus steudneri Schweinf (locally known by the name Gora), a scandent shrub and is characterized by deeply furrowed stems covered with stellate hairs or sometimes with prickles belonging to the family Rosaceae is one of the important edible plants in Ethiopia. The fruits are edible in nature and the plant is utilized as a source of food and medicine [9-11]. It is reported that different parts of the plant namely leaf, stem and fruit are consumed by gorillas in Bwindi Impenetrable National Park, Uganda [12]. In an earlier study, we reported antioxidant [13] and antidiabetic [14] potential of leaves and the ripe and unripe fruits of R. steudneri [15].
Butyl isobutyl phthalate (BIP) (fig.1) is a phytochemical reported rarely from higher plants. BIPwas found to act as a concentration dependent, non-competitive inhibitory activity against α-glucosidase [16,17], antimutagenic [18,19], cytotoxic [20], antifungal [21] and antibacterial [19] activity. However, the information about the interaction of BIP in the catalytic active pocket of α-glucosidase is scanty or limited. In that regard, the current study carried out by us aims at in vitro and in silico investigation of α-glucosidase inhibitory activity of BIP isolated from chloroform fraction of R. steudneri leaves as a possible therapy for type 2 diabetes mellitus.
Materials and Methods
Collection and identification of the plant
The matured R. steudneri leaf material was collected from Nekemte (9°5′N 36°33′E to 9.083°N 36.550°E; 2,123 meters above sea level), East Wollega Zone, Oromia Region, Ethiopia during December 2015 and identified by Dr. Tesfaye Awas, Plant Taxonomist, Ethiopian Biodiversity Institute, Addis Ababa, Ethiopia. The R. steudneri specimen was authenticated based on its characteristics under the voucher no: 1983 at the Ethiopian Biodiversity Institute, Addis Ababa, Ethiopia.
Extraction and characterization
The shade-dried leaf powder (1 kg) was extracted with 3 l of 100 % v/v ethanol for 72 h. The filtrate (vacuum filtered) was concentrated using R-120 rotavap. The percentage yield of concentrated mass was found to be 27.5 % w/w. BIP was isolated as a pure fraction from the chloroform fraction of ethanol extract of R. steudneri leaves by flash chromatography. BIP was characterized on the basis of data obtained from GC-MS analyses and the results revealed 278.1 m/z for chloroform fraction which is consistent with a BIP structure [22].
In vitro α-glucosidase (EC 3.2.1.20) inhibition assay
The α-glucosidase inhibition assay was performed as described by Tomoki Ohta [23] with necessary modifications. In brief, a pre-incubation mixture of 300 μl containing 250 μl of 80 mM phosphate buffer pH 7.0/Acarbose (0.5, 1 and 2 µg/ml)/BIP (3, 6, 12 and 24 µg/ml) and 50 μl of the enzyme was mixed thoroughly and pre-incubated at 37° for 30 min. Following pre- incubation 500 μl substrate (37 mM Sucrose) was added and again incubated at 37° for 20 min. The reaction was arrested by keeping it in boiling water bath for two min and then cooled for some time. A 250 µl of glucose reagent was added to 50 µl of the reaction mixture and mixed thoroughly. The absorbance was measured at 510 nm (Molecular Devices, Versamax Microplate Reader) upon incubating the reaction mixture at 25° for 10 min. The percent of inhibition was calculated using the formula, Inhibition(%)=(A0-A1)/A0×100, where, A0 is the absorbance of the control and A1 is the absorbance of the sample. The Half-maximal inhibitory concentration (IC50) value was estimated from the percentage inhibition versus concentration plot using non-linear regression formula.
Molecular docking studies
The crystal structure of human α-glucosidase protein, the known target for BIP to treat type 2 diabetes (PDB ID: 1OBB, resolution 1.90 Å) was retrieved from the protein data bank and prepared using the Protein Preparation Wizard [24] available in Schrödinger suite 2018-1. Crystallographic water molecules (water molecules without 3 H bonds) were deleted and hydrogen bonds corresponding to pH 7.0 were added considering the appropriate ionization states for both the acidic and basic amino acid residues. Missing side chain atoms were added and breaks present in the structure were built using Prime (v3.7, Schrödinger 2018-1). Using the OPLS_2005 force field [25] energy of crystal structure was minimized. The active site was defined with a 10 Å radius around the co-crystal ligand (maltose) present in the crystal structure and a grid box was generated at the centroid of the active site for docking. To test the docking parameters low energy conformations BIP was docked into the catalytic pocket of the α-glucosidase using Grid-based Ligand Docking with Energetics (Glide v7.8, Schrödinger 2018-1) in ‘extra precision’ mode without applying any constraints [26]. The final best-docked structure was selected using a Glide score function, Glide energy and Glide Emodel energy. Finally, the lowest-energy docked complexes of BIP/α-glucosidase and maltose/ α-glucosidase were selected for further study.
Molecular dynamics (MD) simulation
The molecular dynamics of the docked complexes of BIP/α-glucosidase and Maltose/α-glucosidase were studied using the AMBER force field in an explicit solvent of water within the Cresset flare software for the MD simulations. The molecular system was solvated with water molecules in an orthorhombic box allowing for a 10 Å buffer region between protein atoms and box sides. Then the system temperature 300°K was run for 10 ns. Data were collected every 100 ps during MD run. 3-D structures and trajectories were visually inspected using the Flare graphical interface [27,28].
Results and Discussion
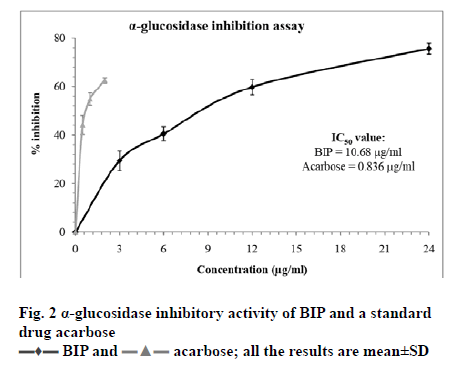
Diabetes mellitus is a metabolic disorder characterized by an elevated level of blood sugar level (postprandial) hyperglycemia. Type 2 diabetes mellitus is the more common type of diabetes mellitus and often leads to serious health complications such as cardiovascular diseases, neuropathy, nephropathy and retinopathy resulting in reduced quality of life and a greater number of deaths worldwide [29,30]. α-amylase and α-glucosidase are the two important carbohydrate digesting enzymes. Postprandial hyperglycemia is often managed by targeting the activity of the key carbohydrate digesting enzyme α-glucosidase in the digestive tract, thereby reducing the blood sugar level and preventing the absorption of sugar [29]. Drugs such as acarbose and miglitol are frequently used to manage postprandial hyperglycemia. However, their use is often associated with side effects. Hence, there is need for developing safe alternatives, especially from natural sources such as plants, with low or no adverse health effects [29,31]. In this study, we have subjected the compound BIP isolated from leaves of R. steudneri for possible inhibitory activity against the enzyme α-glucosidase. It is observed that the compound BIP was effective against inhibiting the activity of α-glucosidase. Fig. 2 depicts the result of α-glucosidase inhibition by different concentrations of BIP (3-24 µg/ml). BIP was found to have inhibitory effects on α-glucosidase in a dose dependent manner with an IC50 was found to be 10.68 µg/ml. At 24 µg/ml concentration, BIP caused 75.61±2.28 % inhibition of α-glucosidase. The reference drug Acarbose was more effective in causing inhibition of α-glucosidase and the IC50 value was found to be 0.836 µg/ml. At concentration 2 µg/ml, Acarbose caused 62.45±1.15 % inhibition of α-glucosidase. To the best of our knowledge, there is scarce information available on the effect of BIP from natural products on the activity of α-glucosidase. Our result is on par with the observation made by Bu et al. [16] and Liu et al. [17] in which BIP isolated from a sea weed Laminaria japonica exhibited inhibitory activity against α-glucosidase with an IC50 value to 38 μM.
The aim of the molecular docking study was to elucidate how BIP bind to the target α-glucosidase,which is essential to catalyze the breakdown of maltose into glucose units. In that regard, the docking results provided appropriate information about the binding affinity, binding energy and orientation of ligandenzyme and substrate-enzyme interactions to inhibit the function of α-glucosidase. The molecular docking was carried out both substrate (maltose) and inhibitor (BIP) and their interaction of ligands with protein was analyzed. The docking protocol was validated using the reported structure of α-glucosidase bound to its substrate maltose. The root mean square (RMS) deviation between the actual and the predicted pose was 0.170 Å, which was well within the acceptable limit of 2.0 Å [32].
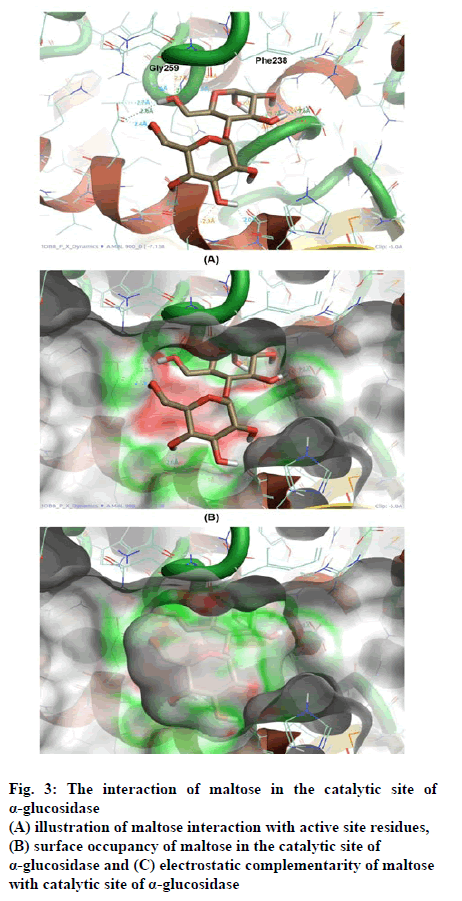
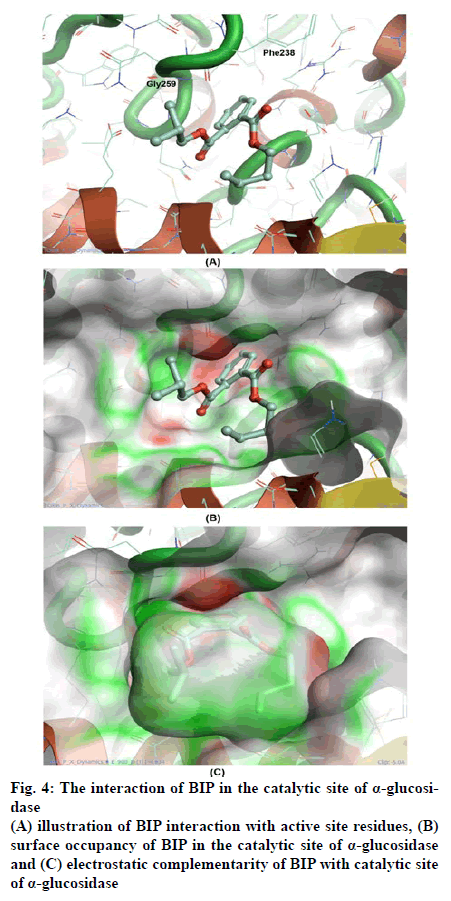
The scores of docking studies are shown in the Table 1, from the total-scores, the IC50 of BIP in touch with the glide score. Docking study revealed that interactions were hydrogen bond and aromaticity. The interactions were dominated in the region of Gly259 to A Phe238 amino acid residues content due to pronouncing existence of the catalytic site in the region as shown in fig. 3A and 4A.
| Compound | Docking score (kcal/mol) | Bond forming atom |
|---|---|---|
| BIP | ̶ 6.3 | A Gly259 H (H-Bond) A Phe238 CG (π−π stacking) |
| Maltose | ̶ 6.9 | A Gly259 H (H-Bond) A Phe238 O (H-Bond) A Ans 239 HD22 (H-Bond) |
Table 1: Docking Score and Interaction Residues for the Binding of Bip and Maltose with Alpha-Glucosidase
Figure 3: The interaction of maltose in the catalytic site of
α-glucosidase
(A) illustration of maltose interaction with active site residues,
(B) surface occupancy of maltose in the catalytic site of
α-glucosidase and (C) electrostatic complementarity of maltose
with catalytic site of α-glucosidase
The interaction of maltose was facilitated by three major contacts formed by hydrogen bonds, whereas the interaction of BIP was mediated by hydrogen bond and π–π stacking each. Though the glide score of both the molecules are significantly similar, the interaction pattern is differed because of varied chemistry of both the molecules. Maltose being the highly polar molecule possessed comparatively high h-bonds with an enzyme. However, the interaction of BIP is also sufficient to possess significant inhibition. The occupancy and conformation of aromatic ring in both substrate and inhibitor was seen similar as shown in fig. 3B and 4B.
The electrostatic complementarity (EC) of BIP shown in fig. 4C found better than the EC of maltose shown in fig. 3C. Inclusion of EC significantly improves the docking results for most systems by ranking the nearly correct solutions as highly probable. In that concern the EC analysis has described the substantial similarity between substrate and inhibitors in interaction with α-glucosidase. However, it requires advanced computational calculations like MD simulations, to illustrate the stability in protein-ligand interaction.
Molecular dynamics (MD) simulation was conducted using docked conformation of BIP/α-glucosidase and Maltose/α-glucosidase complexes to explore the dynamic stability of system.MD simulation calculations performed using OpenMM, to study the stability of protein-ligand complexes. The interactive plots derived for (potential energy, temperature, box volume and density), have simplified the understanding of system stability.
The system was simulated for 50 ps for equilibration. The final simulation run was for a total of 10 ns, during which 1000 structures enumerated were saved in the trajectory. To understand the stability of the complex during MD simulation, the protein backbone frames were aligned to the backbone of initial frame and then the RMS deviation was calculated with respect to the initial frame. The RMS deviations between the original structure and the structure enumerated during MD simulation were plotted as shown in fig. 5A. The protein backbone RMS deviation recorded during simulation shows large deviation for the initial 100 ps due to the initial protein structural stabilization; after that showed a steady state dynamics. The backbone structural deviations observed for the latter phase of 1 to 10 ns was in the range of 1.4 Å to 1.8 Å compared to that of original structure. From 1 ns to till the end of simulation, the total RMS deviation of protein was within a range of 0.4 Å. The total potential energy, density, temperature and box volumeof the system were maintained at -170398.84 kcal/mole, 1.034 g/ml, 295.97 K and 561.06 nm^3 respectively as shown in fig. 5B-E. This clearly suggests that 10 ns of simulation are sufficient for stabilizing these complexes and the maltose/α-glucosidase complex is stable.
Similarly, The RMS deviations between the original BIP/α-glucosidase structure and the structure enumerated during MD simulation were plotted as shown in fig. 6A. The protein backbone RMS deviation recorded during simulation shows large deviation for the initial 2.5 ns due to the initial protein structural stabilization; after that found to gain the stability. The backbone structural deviations observed for the latter phase of 2.5 to 10 ns was in the range of 1.4 Å to 2.0 Å compared to that of original structure. From 2.5 ns to till the end of simulation, the total RMS deviation of protein was within a range of 0.6 Å. The total potential energy, density, temperature and box Volume of the system were maintained at -170595.25 kcal/mole, 1.033 g/ml, 295.48 K and 561.84 nm^3 respectively as shown in fig. 6B-E. This clearly suggests that 10 ns of simulation are sufficient for stabilizing these complexes and the BIP/α-glucosidase complex is stable.
Comparatively, no is significant difference in MD sim- ulation was seen. However, the total potential energy of BIP/α-glucosidase complex was better than Maltose/α-glucosidase. Hence, BIP can be a potential enzyme in- hibitor of α-glucosidase. We present the possible mode of action of the phytochemical BIP from chloroform fraction of R. steudneri which can be a potential clini- cal intervention for type 2 diabetes which acts through α-glucosidase inhibition. We have established the phar- macological activity both by in vitro enzyme activity and in silico ligand binding activity through molecular docking analysis. Since both the studies reiterates about the potency of the ligand to inhibit the α-glucosidase along with highly favorable rule of 5 statistics, this li- gand can further be taken for drug development pro- cess.
References
- Gerich JE. Clinical significance, pathogenesis and management of postprandial hyperglycemia.Arch Intern Med2003;163(11):1306-16.
- Tibaldi J. Importance of postprandial glucose levels as a target for glycemic control in type 2 diabetes.South Med J2009;102(1):60-6.
- Woerle HJ, Neumann C, Zschau S, Tenner S, Irsigler A, Schirra J, et al. Impact of fasting and postprandial glycemia on overall glycemic control in type 2 diabetes. Importance of postprandial Glycemia to achieve target HbA1clevels. Diabetes Res Clin Pract 2007;77(2):280-5.
- Node K, Inoue T. Postprandial hyperglycemia as an etiological factor in vascular failure.Cardiovasc Diabetol 2009;8:23.
- Joshi SR, Standl E, Tong N, Shah P, Kalra S, Rathod R. Therapeutic potential of α-glucosidase inhibitors in type 2 diabetes mellitus: an evidence-based review.Expert Opin Pharmacother 2015;16(13):1959-81.
- Van de Laar FA. Alpha-glucosidase inhibitors in the early treatment of type 2 diabetes.Vasc Health Risk Manag 2008;4(6):1189-95.
- Van de Laar FA, Lucassen PLBJ, Akkermans RP, Van de Lisdonk EH, Rutten GEHM, Van Weel C. Alpha-glucosidase inhibitors for type 2 diabetes mellitus.Cochrane Database Syst Rev 2005;18(2):CD003639.
- Mata R,Cristians S,Escandón-Rivera S,Juarez-Reyes K, Rivero-Cruz I. Mexican antidiabetic herbs: valuable sources of inhibitors ofα-glucosidases. J Nat Prod 2013;76:468-83.
- Giday M, Asfaw Z, Woldu Z. Medicinal plants of the Meinit ethnic group of Ethiopia: An ethnobotanical study. J Ethnopharmacol 2009;124:513-21.
- Yineger H, Kelbessa E, Bekele T, Lulekal E. Plants used in traditional management of human ailments at Bale Mountains National Park, Southeastern Ethiopia. J Med Plants Res 2013;2:132-53.
- Kefalew A, Asfaw Z, Kelbessa E. Ethnobotany of medicinal plants in Ada’a District, East Shewa Zone of Oromia Regional State, Ethiopia. J Ethnobiol Ethnomed 2015;11:25.
- Rothman JM, Dierenfeld ES, Molina DO, Shaw AV, Hintz HF, Pell AN. Nutritional chemistry of foods eaten by gorillas in Bwindi Impenetrable National Park, Uganda. Am J Primatol 2006;68:675-91.
- Raghavendra HL, Kekuda PTR. Preliminary phytochemical analysis, antiradical and lipid peroxidation inhibitory activity of Rubus steudneri Schweinf. (Rosaceae). Int J Green Pharm 2018;12:49-55.
- Raghavendra HL, Upashe SP, Reyes DRA, Floriano JF. Antidiabetic and antioxidant activity of Rubus apetalus Poir. and Rubus steudneri Schweinf. leaf extract on alloxan induced diabetes mellitus. J Bioanal Biomed 2019;1:149-54.
- Raghavendra HL, Kekuda PTR. Antiradical and lipid peroxidation inhibitory activity of ripe and unripe fruit of Rubus steudneri Schweinf. (Rosaceae). Pharmacog J 2018;10:818-22.
- Bu T, Liu M, Zheng L, Guo Y, Lin X. α-glucosidase inhibition and thein vivohypoglycemic effect of butyl-isobutyl-phthalate derived from theLaminaria japonicarhizoid. Phytother Res 2010;24:1588-91.
- Liu M, Zhang W, Qiu L, Lin XK. Synthesis of butyl-isobutyl-phthalate and its interaction with α-glucosidase in vitro. J Biochem 2011;149:27-33.
- Cruz-Ramírez SG, López-Saiz CM, Plascencia-Jatomea M, MachiLara L, Rocha-Alonzo F, Márquez-Ríos E et al. Isolation and identification of an antimutagenic phthalate derivative compound from octopus (Paractopus limacualtus). Trop J Pharm Res 2015;14: 1257-67.
- Hussain A, Rather MA, Dar MS, Aga MA, Ahmad N, Manzoor A et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorganic Med Chem Lett 2017;27:2579-82.
- Zeng X, Xiao X, Li D, Gu Q, Wang F. Isolation, identification and screening of microorganisms for cytotoxic activities from deep sea sediments at different pacific stations. World J Microbiol Biotechnol 2010;26:2141-50.
- Tian JJ, Gao XX, Zhang WM, Wang L, Qu LH. Molecular identification of endophytic fungi from Aquilaria sinensis and artificial agarwood induced by pinholes-infusion technique. Afr J Biotechnol 2013;12:3115-31.
- Hallur RLS, Motamarri CVNL, Ramamoorthy PKT, Murthy CD, Siddappa RPH, Bramhanakonda VN. GC-MS fingerprint and in vitro cytotoxic studies of Rubus steudnerileaf fractions against MCF-7 breast cancer cell line. Pharmacogn Mag 2020;16(72):44.
- Tomoki O, Shigefumi A, Tadashi O, Shuji Y, Hideyuki K. Alpha-glucosidase inhibitory activityof a 70 % methanol extract from ezoishige (Pelvetia babingtonii de Toni) and its effect on the elevation of blood glucose level in rats. Biosci Biotechnol Biochem 2002;66:1552-4.
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 2013;27:221-34.
- Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 2010;6:1509-19.
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 2006;49:6177-96.
- Guo Z, Mohanty U, Noehre J, Sawyer TK, Sherman W, Krilov G. Probing the α-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem Biol Drug Des 2010;75:348-59.
- Bharath BR, Damle H, Ganju S, Damle L. In silico screening of known small molecules to bind ACE2 specific RBD on Spike glycoprotein of SARS-CoV-2 for repurposing against COVID-19. F1000 Research 2020;9:663.
- Mahayasih PGM, Elya B, Hanafi M. Alpha-glucosidase inhibitory activity of Garcinia lateriflora Blume leaves. J App Pharm Sci 2017;7:100-4.
- Alqahtani AS, Hidayathulla S, Rehman MT, ElGamal AA, Al-Massarani S, Razmovski-Naumovski V, et al. Alpha-amylase and alpha-glucosidase enzyme inhibition and antioxidant potential of 3-Oxolupenal and Katononic acid isolated from Nuxia oppositifolia. Biomolecules 2020;10:61.
- Laoufi H, Benariba N, Adjdir S, Djaziri R. In vitro α-amylase and α-glucosidase inhibitory activity of Ononis angustissima extracts. J App Pharm Sci 2017;7:191-8.
- Kontoyianni M, McClellan LM, Sokol GN. Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 2004;47:558-65.
 acarbose; all the results are mean±SD
acarbose; all the results are mean±SD