- *Corresponding Author:
- Qiquan Liu
Department of Gastroenterology, First Affiliated Hospital of Hebei University of Chinese Medicine, Shijiazhuang, Hebei 050011, China
E-mail: liuqq56@163.com
| This article was originally published in a special issue, “Innovations in Biomedical Research and Drug Development” |
| Indian J Pharm Sci 2023:85(3) Spl Issue “72-82” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The aim of this study was to identify the potential compounds, core targets and possible mechanism of Huanglian Jiedu decoction in treating ulcerative colitis based on network pharmacology and molecular docking. At first, potential compounds of Huanglian Jiedu decoction were retrieved from traditional Chinese medicine systems pharmacology. And then, the targets related to compounds and ulcerative colitis was obtained from traditional Chinese medicine systems pharmacology, Online Mendelian Inheritance in Man, GeneCards and DisGeNET. Next, Cytoscape was used to visualize drug-compound-common target-disease network and protein-protein interaction network. Moreover, gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis was performed by database for annotation, visualization and integrated discovery to investigate possible mechanism of Huanglian Jiedu decoction against ulcerative colitis. At last, molecular docking verified the reliability of the prediction results. 55 compounds and 84 targets of Huanglian Jiedu decoction were screened out as potential players on ulcerative colitis. After network analyses, 10 core compounds (quercetin, kaempferol, wogonin, baicalein, acacetin, 5-hydroxy-7-methoxy-2-(3,4,5- trimethoxyphenyl) chromone, beta-sitosterol, moslosooflavone, 5,7,4'-trihydroxy-8-methoxyflavone, oroxylin A) and 10 core targets (interleukin-6, interleukin-1 beta, tumor necrosis factor-alpha, threonine-protein kinases, tumor antigen p53, prostaglandin-endoperoxide synthase 2, JUN, CXCL8, C-C motif chemokine 2, matrix metalloproteinase-9) were identified. Furthermore, the inflammatory response, tumor necrosis factoralpha signaling pathway, pathways in cancer, T cell receptor signaling pathway, toll-like receptor signaling pathway and nuclear factor kappa B signaling pathway may be involved in the treatment of ulcerative colitis using Huanglian Jiedu decoction. This study reveals that Huanglian Jiedu decoction contains multiple ingredients, multiple targets and multiple pathways in treating ulcerative colitis, which provides a basis for further research.
Keywords
Ulcerative colitis, inflammatory bowel disease, nuclear factor kappa B, matrix metalloproteinase-9, diarrhea
Ulcerative Colitis (UC), as a chronic idiopathic Inflammatory Bowel Disease (IBD), is characterized by long-lasting mucosal inflammation of the colon[1,2]. The main symptoms of UC are abdominal pain, bloody diarrhea and tenesmus. With the increasing prevalence of IBD, UC evidently influences the health and life quality of patients all over the world. The incidence of UC is higher in developed countries than other regions of the world. However, the incidence and prevalence of UC in Asian countries is increasing year by year[3]. In China, a previous meta-analysis of IBD patients based on population and hospital studies showed that the incidence rate of UC was 1.2/100 000 persons[4]. At present, the pharmacological management of UC is focused on controlling symptoms with 5-Aminosalicylates (5- ASA), corticosteroids and thiopurines, which is still not ideal[5]. Given this, discovering more effective and less toxic treatments for UC is ur gently needed.
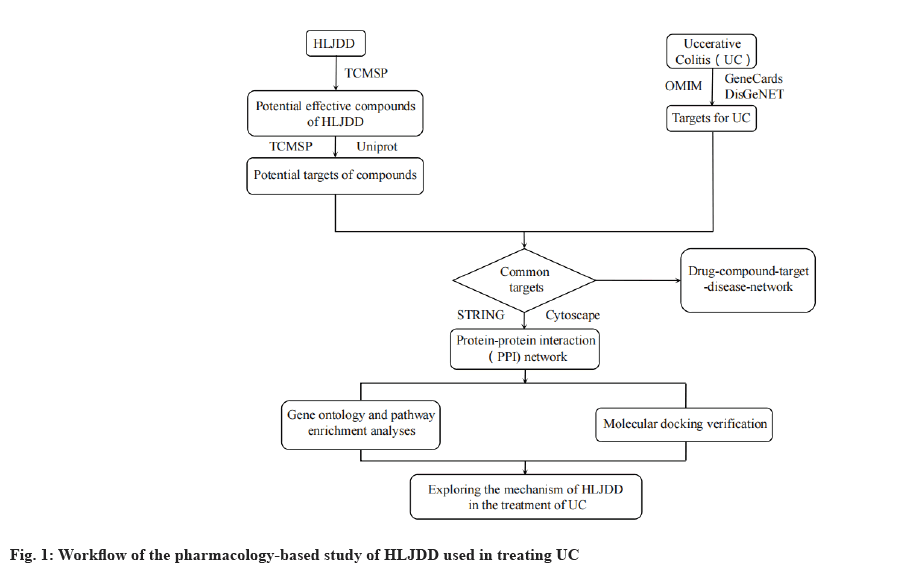
Traditional Chinese Medicine (TCM), as a significant part of complementary and alternative medicine system, has been used to treat and prevent UC for a long time[6]. TCM can alleviate the symptoms of UC, heal the ulcers and improve the quality of life. Hence, it is well worth studying in TCM prescription against UC deeply. Huanglian Jiedu Decoction (HLJDD) originates from “The Handbook of Prescriptions for Emergencies”, is an important and classic TCM formula to clear heat and detoxify. HLJDD is composed of four herbs; Coptidis rhizoma (Huanglian (HL)), Scutellariae radix (Huangqin (HQ)), Phellodendri chinensis cortex (Huangbai (HB)) and Gardeniae fructus (Zhizi (ZZ)) at a ratio of 3:2:2:3, respectively. At present, HLJDD has been widely used in treating inflammatory gastrointestinal disease including UC[7]. Based on the TCM theory, the pathogenesis of UC is closely associated with toxic heat, which could be solved by HLJDD. Modern pharmacological study has shown that the n-butanol fraction of HLJDD can significantly control the symptoms and pathological damage of UC mice and inhabit the inflammatory response[8]. Chinese researchers have also reported that HLJDD could alleviate the colonic mucosal inflammation and damage in UC mice by reducing the levels of Tumor Necrosis Factor alpha (TNF-α), Interleukin (IL)-6 and IL-1 beta (β)[9]. However, the clear molecular mechanism of HLJDD against UC still lacks systematic understanding and enough relevant studies. Network pharmacology, first proposed by Andrew L Hopkins, has been a powerful tool to illuminate our understanding of drug action[10]. Similar to TCM, network pharmacology is also holistic and systematic in methodology, which can be applied to TCM herbal studies[11]. Based on network pharmacology, we can discover new bioactive ingredients and special biomarkers, elucidating possible mechanisms of interactive actions between multicompounds and multitargets of herb formula in TCM[12]. In this study, we employed network pharmacology technology to predict the mechanism of HLJDD on UC and the prediction results were verified by molecular docking subsequently. The detailed workflow of our study was exhibited in fig. 1.
Materials and Methods
Collection and screening of potential bioactive compounds in HLJDD:
The collection of four candidate herbs in HLJDD was retrieved by TCM Systems Pharmacology Database and Analysis Platform[13] (TCMSP, https://tcmsp-e.com/), a TCM systems pharmacology platform which contains the relationship between drugs, targets and disorders. According to relevant literature report, the pharmacokinetic properties including Absorption, Distribution, Metabolism and Excretion (ADME) were generally employed to screened out potential bioactive compounds in herbs[14]. As suggested by TCMSP, the compounds whose Oral Bioavailability (OB) ≥30 % and drug-likeness ≥0.18 at the same time could be identified as the potential bioactive compounds in HLJDD for further analysis.
Target fishing for HLJDD:
The compounds in HLJDD depend on their targets to exert their role. After searching in TCMSP with the name of potential compounds in HLJDD, the targets of compounds were obtained. Target names were then standardized through the UniProt database (http://www.uniprot.org/), an informative and resourceful protein knowledgebase[15] and we selected “Homo sapiens” as the potential target species of the potential compounds of HLJDD.
Search targets of UC:
In this process, the key word “UC” was searched in Online Mendelian Inheritance in Man (OMIM)[16] (https://www.omim.org/), GeneCards[17] (https://www.genecards.org/) and DisGeNET[18] (https://www.disgenet.org/) to collect the targets related to UC. UC targets were finally obtained after removing repetitive data. Common targets of both potential bioactive compounds and UC were considered as the related targets of HLJDD treatment for UC. The relationship between UC targets and HLJDD targets was shown by Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Protein-Protein Interaction (PPI) data:
Based on the above research, intersective targets about HLJDD on UC were imported into the STRING platform[19] (https://string-db.org,ver 11.0) to obtain the PPI data. The species was limited to “Homo sapiens”, the confidence score >0.4 as the threshold and the discrete targets were deleted.
Network construction:
Network construction was visualized by Cytoscape software[20] (version 3.8.2) as follows; drug-compound-common target-disease network; PPI network was constructed by linking common targets between UC and HLJDD. Via the network analyzer tool in Cytoscape, the potential core targets of the PPI network were obtained by the rank of degree numbers.
Gene Ontology (GO) and pathway enrichment analysis:
To figure out the possible mechanisms of HLJDD on UC, intersective targets were imported into the Database for Annotation, Visualization and Integrated Discovery[21] (DAVID), (https://david. ncifcrf.gov/) with species limited to “Homo sapiens”. DAVID is a functional annotation tool to predict possible Cellular Component (CC), Molecular Function (MF), Biological Process (BP) and signaling pathway associated with specific genes. With p<0.01 to keep the reliability of the enrichment analyses and the analytic results were visualized by R language.
Molecular docking:
We used molecular docking technology to verify the reliability of the network pharmacology prediction results. The structures of core compounds in HLJDD were downloaded from the PubChem and the Three- Dimensional (3D) structures of the core target proteins were obtained from the Protein Data Bank (PDB)[22] (https://www.rcsb.org/). The molecular docking and conformation scoring were carried out in the AutoDock Vina and the heat map was drawn by the GraphPad Prism 8. The Pymol and Maestro 11.9 were used to draw the structure of the representative docking results.
Results and Discussion
A total of 80 compounds were screened out and collected from TCMSP based on the screening criteria; 25 in HB, 11 in HL, 32 in HQ and 12 in ZZ, respectively; among them, 14 compounds were duplicated and removed. For example, beta-sitosterol existed in HB, HQ and ZZ. By using TCMSP, 228 targets of potential bioactive compounds in HLJDD were obtained after deleting duplicates.
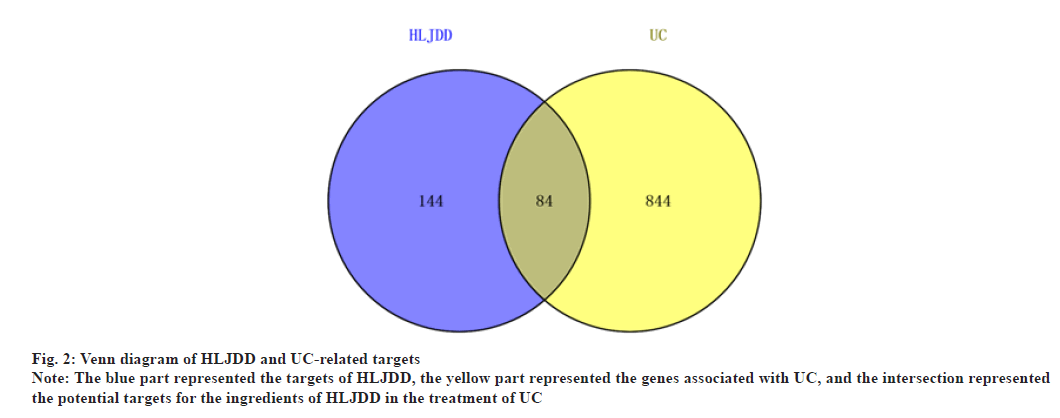
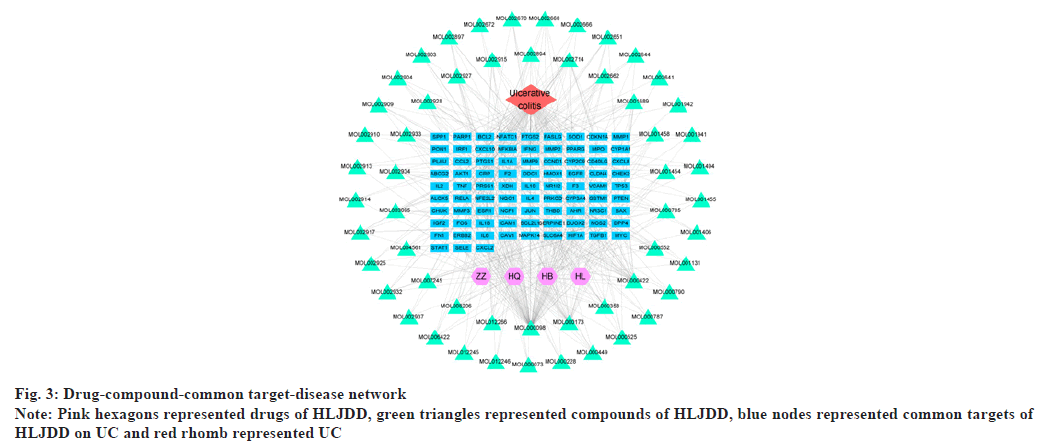
With “UC” as the key word, a combined total of 928 disease targets were collected from OMIM, GeneCards and DisGeNET databases after removing duplicates. As a result, the intersections of predicted targets of HLJDD and disease targets resulted in 84 common targets for UC (fig. 2). At last, 55 potential compounds were identified from HLJDD and some bioactive compounds were deleted in lack of common targets (Table 1). By using Cytoscape, we constructed a network relationship among HLJDD compounds and common targets, the resulting network included 144 nodes and 508 interaction edges (fig. 3). The degree values of compounds were then obtained. Quercetin has 76 potential targets, followed by kaempferol with 27 and wogonin with 24. Top 10 compounds were considered to be involved in the treatment of UC by HLJDD.
| Molecule ID | Molecule Name | OB (%) | DL | HERB |
|---|---|---|---|---|
| MOL001454 | Berberine | 36.86 | 0.78 | HL, HB |
| MOL002894 | Berberrubine | 35.74 | 0.73 | HL, HB |
| MOL002897 | Epiberberine | 43.09 | 0.78 | HL, HQ |
| MOL002903 | (R)-Canadine | 55.37 | 0.77 | HL |
| MOL002904 | Berlambine | 36.68 | 0.82 | HL |
| MOL000785 | Palmatine | 64.6 | 0.65 | HL, HB |
| MOL000098 | Quercetin | 46.43 | 0.28 | HL, HB, ZZ |
| MOL001458 | Coptisine | 30.67 | 0.86 | HL, HQ, HB |
| MOL002668 | Worenine | 45.83 | 0.87 | HL, HB |
| MOL001689 | Acacetin | 34.97 | 0.24 | HQ |
| MOL000173 | Wogonin | 30.68 | 0.23 | HQ |
| MOL000228 | (2R)-7-hydroxy-5-methoxy-2-phenylchroman-4-one | 55.23 | 0.2 | HQ |
| MOL002714 | Baicalein | 33.52 | 0.21 | HQ |
| MOL002909 | 5,7,2,5-Tetrahydroxy-8,6-dimethoxyflavone | 33.82 | 0.45 | HQ |
| MOL002910 | Carthamidin | 41.15 | 0.24 | HQ |
| MOL002913 | Dihydrobaicalin_qt | 40.04 | 0.21 | HQ |
| MOL002914 | Eriodyctiol (flavanone) | 41.35 | 0.24 | HQ |
| MOL002915 | Salvigenin | 49.07 | 0.33 | HQ |
| MOL002917 | 5,2',6'-Trihydroxy-7,8-dimethoxyflavone | 45.05 | 0.33 | HQ |
| MOL002925 | 5,7,2',6'-Tetrahydroxyflavone | 37.01 | 0.24 | HQ |
| MOL002927 | Skullcapflavone II | 69.51 | 0.44 | HQ |
| MOL002928 | Oroxylin A | 41.37 | 0.23 | HQ |
| MOL002932 | Panicolin | 76.26 | 0.29 | HQ |
| MOL002933 | 5,7,4'-Trihydroxy-8-methoxyflavone | 36.56 | 0.27 | HQ |
| MOL002934 | Neobaicalein | 104.34 | 0.44 | HQ |
| MOL002937 | Dihydrooroxylin | 66.06 | 0.23 | HQ |
| MOL000358 | Beta-sitosterol | 36.91 | 0.75 | HQ, HB, ZZ |
| MOL000525 | Norwogonin | 39.4 | 0.21 | HQ |
| MOL000552 | 5,2'-Dihydroxy-6,7,8-trimethoxyflavone | 31.71 | 0.35 | HQ |
| MOL000073 | Ent-Epicatechin | 48.96 | 0.24 | HQ |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | HQ, HB, ZZ |
| MOL008206 | Moslosooflavone | 44.09 | 0.25 | HQ |
| MOL012245 | 5,7,4'-Trihydroxy-6-methoxyflavanone | 36.63 | 0.27 | HQ |
| MOL012246 | 5,7,4'-Trihydroxy-8-methoxyflavanone | 74.24 | 0.26 | HQ |
| MOL012266 | Rivularin | 37.94 | 0.37 | HQ |
| MOL002641 | Phellavin_qt | 35.86 | 0.44 | HB |
| MOL002644 | Phellopterin | 40.19 | 0.28 | HB |
| MOL002651 | Dehydrotanshinone II A | 43.76 | 0.4 | HB |
| MOL002662 | Rutaecarpine | 40.3 | 0.6 | HB |
| MOL002666 | Chelerythrine | 34.18 | 0.78 | HB |
| MOL002670 | Cavidine | 35.64 | 0.81 | HB |
| MOL002672 | Hericenone H | 39 | 0.63 | HB |
| MOL000787 | Fumarine | 59.26 | 0.83 | HB |
| MOL000790 | Isocorypalmine | 35.77 | 0.59 | HB |
| MOL001131 | Phellamurin_qt | 56.6 | 0.39 | HB |
| MOL001455 | (S)-Canadine | 53.83 | 0.77 | HB |
| MOL006422 | Thalifendine | 44.41 | 0.73 | HB |
| MOL001406 | Crocetin | 35.3 | 0.26 | ZZ |
| MOL001941 | Ammidin | 34.55 | 0.22 | ZZ |
| MOL004561 | Sudan III | 84.07 | 0.59 | ZZ |
| MOL000422 | Kaempferol | 41.88 | 0.24 | ZZ |
| MOL001494 | Mandenol | 42 | 0.19 | ZZ |
| MOL001942 | Isoimperatorin | 45.46 | 0.23 | ZZ |
| MOL003095 | 5-Hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl) chromone | 51.96 | 0.41 | ZZ |
| MOL007245 | 3-Methylkempferol | 60.16 | 0.26 | ZZ |
Table 1: Potential Effective Compounds of HLJDD
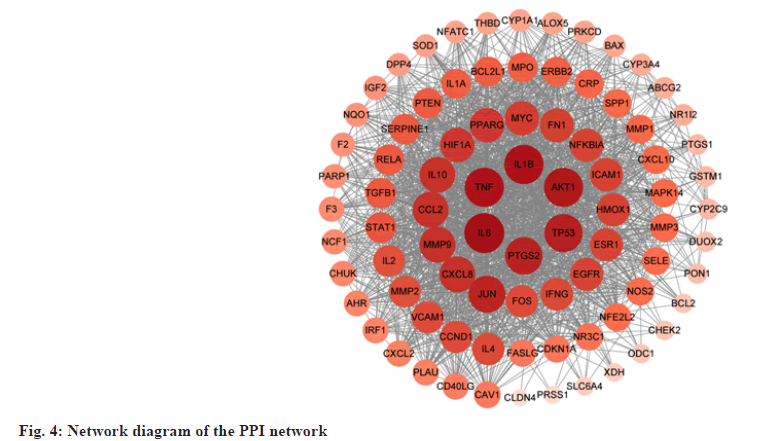
PPI data were obtained from STRING by uploading 84 common targets and PPI network was mapped by Cytoscape, consisting of 84 nodes and 1568 interaction edges. In the network, the greater the degree, the larger and darker color the node was (fig. 4). 10 targets with the largest degree value (degree ≥62) were selected as core targets for UC. The core targets, which may play a significant role against UC, were (IL-6, degree=75), (IL-1β, degree=73), (TNF, degree=73), (RAC-alpha serine/Threonine-Protein Kinase (AKT1), degree=72), cellular (Tumor Antigen p53 (TP53, degree=69), Prostaglandin G/H Synthase 2 (PTGS2), degree=67), Transcription Factor AP-1 (JUN, degree=66), IL-8 (CXCL8, degree=62), C-C Motif Chemokine 2 (CCL2, degree=62) and Matrix Metalloproteinase-9 (MMP-9, degree=62).
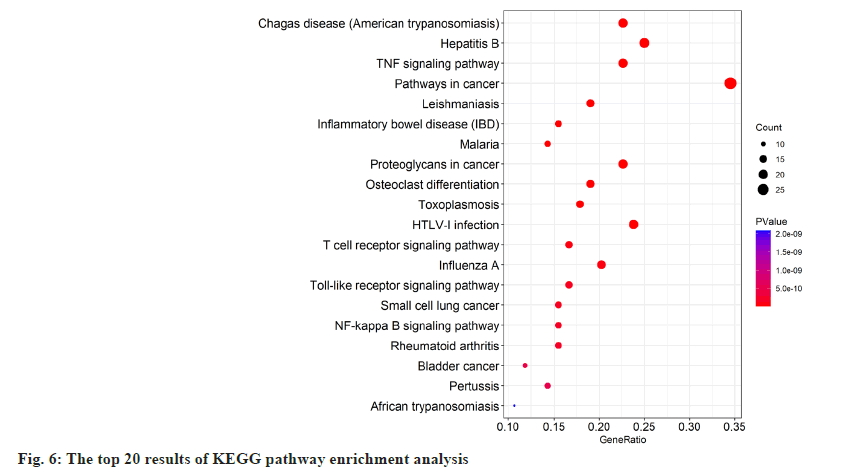
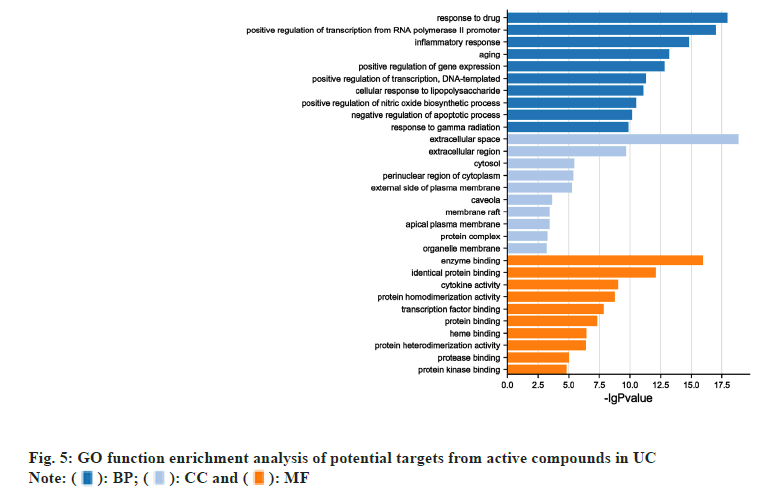
GO term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results were acquired by DAVID. Top 10 enriched conditions in BP, CC and MF were exhibited in fig. 5. 255 BPs, 21 CCs and 42 MFs enriched for common targets have a p<0.01. In GO term enrichment, the BP of HLJDD against UC may relate to response to drug, positive regulation of transcription from Ribonucleic Acid (RNA) polymerase II promoter, inflammatory response and so on. Top 3 MF were enzyme binding, identical protein binding and cytokine activity. Cell compound analysis showed that extracellular space, extracellular region, cytosol, perinuclear region of cytoplasm and external side of plasma membrane were ranked as the top 5 CCs. Besides, 77 KEGG pathways were recognized as p<0.01 and top 20 KEGG pathway’s enrichment analysis was shown in fig. 6. The KEGG analysis results showed that the mechanism of HLJDD against UC might focus on multiple signaling pathways such as TNF signaling pathway, pathways in cancer, T cell receptor signaling pathway, Toll-Like Receptor (TLR4) signaling pathway and Nuclear Factor Kappa B (NF-κB) signaling pathway.
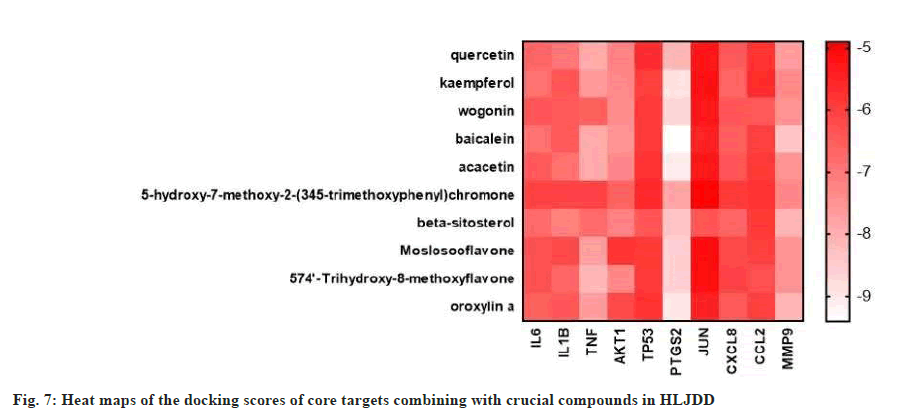
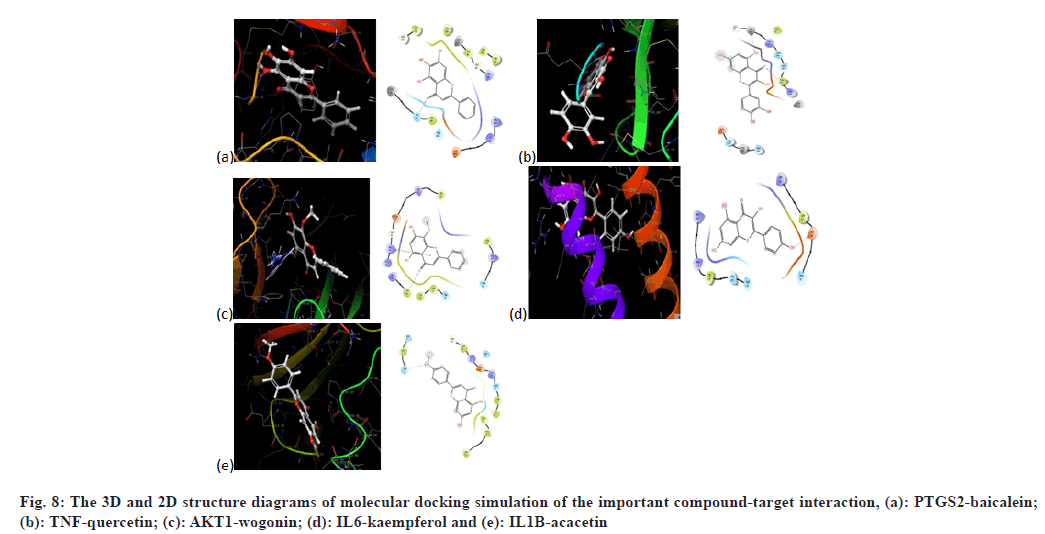
The core targets IL-6, IL-1β, TNF, AKT1, TP53, PTGS2, JUN, CXCL8, CCL2 and MMP-9 were used for molecular docking with the top 10 crucial compounds in HLJDD (fig. 7). Most of the compounds of HLJDD had good binding with core targets and markers, which means that HLJDD has a strong potential used to treat UC. Results showed that baicalein has a strong binding ability with PTGS2 (Vina score=-9.4 kca1/ mol). Quercetin has a strong binding ability with TNF (Vina score=-7.9 kca1/mol). Wogonin binds with AKT1 (Vina score=-7.4 kca1/mol), kaempferol binds with IL-6 (Vina score=-6.9 kca1/mol) and acacetin binds with IL-1β (Vina score=-6.9 kca1/mol). Further molecular docking revealed that baicalein binds with PTGS2 through hydrophobic interaction at sites LEU152, VAL46, CYS47, PRO153 and ARG469 and has hydrogen bonding action with HIS39, TYR130 and CYS47. Quercetin binds with TNF through hydrogen interaction at sites including CYS98, ARG104 and TYR103 and has hydrophobic bonding action with GLN130 and GLU131. Wogonin has hydrogen bonding action with PHE88 and GLY16 by binding with AKT1. Kaempferol binds with IL-6 through hydrogen interaction at ARG30 and has hydrophobic bonding action with LEU33 and LEU178. Acacetin binds with IL-1ß through hydrogen interaction at sites including ASN7, PRO87 and ASN66 and has hydrophobic bonding action with PRO91 and LYS63. Fig. 8 illustrates these local structures of molecular docking in detail.
UC is a type of IBD and it is a great threat to human health. However, there is in lack of relatively effective and safe way to treat this disease. TCM has accumulated rich clinical experience for many years in treating complex disease such as UC. In TCM, UC belongs to the category of “dysentery” and so on. HLJDD is widely used in China to treat UC, but its mechanism of action is still not fully understood. It has been revealed that HLJDD and its effective fraction can alleviate UC by inhibiting Cyclooxygenase-2 (COX-2) protein expression and PLA2, 5-LOX activity to regulate arachidonic acid metabolism and glycerophospholipid metabolism[23]. In this study, a network pharmacological prediction was conducted on the bioactive compounds of the four TCMs (Coptidis rhizoma, Scutellariae radix, Phellodendri chinensis cortex, and Gardeniae fructus) in HLJDD and UC targets. This research could lay the foundation for further experimental verification and provide effective therapeutic strategies for better treatment of UC in the future.
Based on the results of network pharmacology, we speculated that core compounds in HLJDD play significant roles in UC treatment because of their high degree values. Quercetin is known to be a natural dietary flavonoid which has several biological effects including anti-inflammatory, anti-tumor, antioxidant and gastrointestinal cytoprotective effects[24]. Modern research has found that quercetin administration could suppress inflammation in UC-organoids from mice[25]. Researchers also found quercetin-loaded microcapsules could ameliorate the inflammation in colon[26]. Kaempferol, considered being a natural flavonols and its anti-inflammatory property has been confirmed[27]. The plasma levels of Nitric Oxide (NO), Prostaglandin E2 (PGE2) and Leukotriene B4 (LTB4) in Dextran Sodium Sulfate (DSS) induced colitis in C57BL/6J mice can be significantly reduced by oral kaempferol, indicating that kaempferol can be an effective antiinflammatory agent in protecting colonic mucosa from UC[28]. Wogonin is also a kind of flavonoid showing anti-inflammatory potential inhibiting UC[29]. By up-regulating the IL-10 production, wogonin could enhance the therapeutic effects of mesenchymal stem cells in DSS induced colitis[30]. By improving intestinal epithelial barrier via AHR/IL-22 pathway, baicalein can ameliorates UC[31]. Baicalein can regulate Treg cell differentiation and maintain immune homeostasis in DSS induced colitis mice, which might be a potential drug for UC[32]. Acacetin is an O-methylated flavone which possesses anti-inflammatory, anticancer and antiperoxidative activities. Research has shown that acacetin could inhibit the macrophage inflammatory response and adjust intestinal microbiota to alleviate DSS-induced colitis in mice[33]. The results of molecular docking also verified that the core compounds have good binding properties with most core targets. These results could provide novel thought on studying the mechanism of specific compounds in HLJDD against UC. However, some of the compounds are in lack of ample evidences related to UC to support our study’s prediction results, which needs further study to verify.
In order to further explore the possible mechanism of HLJDD on UC, GO and KEGG pathway analysis is indispensable. According to CC analysis results, HLJDD mainly took action in the region of intracellular and extracellular. So far, the pathogenesis of UC has not been elucidated. From top 10 enriched conditions in BP, we could speculate that response to drug, positive regulation of transcription from RNA polymerase II promoter, inflammatory response and negative regulation of apoptotic process were mainly involved in the BP of HLJDD in treating UC. These BP are mainly associated with the process of occurrence and development of UC. KEGG enrichment analysis suggested that HLJDD targeted genes were mainly associated with inflammation related diseases. Furthermore, we found that HLJDD employed a therapeutic effect on UC through multiple pathways. Among them, TNF signaling pathway, pathways in cancer, T cell receptor signaling pathway, TLR4 signaling pathway and NF-κB signaling pathway were the key points and principal signaling pathways that might be related to the mechanism of HLJDD against UC.
PPI analysis revealed that the top 10-ranking genes, IL-6, IL-1ß, TNF, AKT1, TP53, PTGS2, JUN, CXCL8, CCL2 and MMP9 might be the crucial targets regulated by HLJDD for treating UC. Most of them have been documented associated with the above signaling pathways, which confirms the reliability of the KEGG analysis in the other aspect. IL-6, a key proinflammatory cytokine, exists in the pathogenesis of UC and the level of IL-6 is linked to the disease severity[34]. As a central element in the regulation of inflammation at the gastrointestinal mucosal level, IL1B can be served as a key target for IBD prevention and treatment[35]. TP53, a tumor suppressor gene, its mutations is the first step in the evolution from inflamed colonic epithelium to Colorectal Cancer (CRC), demonstrating the close connection between TP53 and UC[36]. According to research data, haplotype of PTGS2 contributes to the susceptibility of IBD, including UC[37]. As an immediate-early gene, proto-oncogene JUN (c-Jun) plays a significant role in inflammatory responses. The expression of c-Jun increases rapidly in cells, enabling cells to adapt to environmental changes when faced with external stimuli[38]. CXCL8 is acting as a proinflammatory chemokine. Compared with healthy volunteers, patients with UC have elevated levels of CXCL8 and CCL2 in the colonic mucosa[39].
Although the pathogenesis of UC has not yet been clearly elucidated, scientists commonly believe that immune system and inflammation bears on it. It has been universally acknowledged that cancer is closely related to inflammation[40]. Longstanding chronic inflammation mainly contributes to the occurrence of CRC[41]. Researchers believe that increased duration and extent of UC increases the risk of CRC[42]. A meta-analysis concerning about 116 studies found that the incidence of CRC positively correlated with the duration of UC[43]. According to the prediction results, HLJDD possibly could treat UC by inhibiting the process from inflammation to cancer, which needs further studies. TNF is a pleiotropic cytokine with important functions in inflammatory disease pathogenesis, such as inducing inflammation, orchestrating the tissue recruitment of immune cells and promoting tissue destruction[44]. Anti-TNF therapy has been proven to be an effective means to induce clinical and endoscopic remission in UC patients; however, its relevant risks require deep concern[45]. T-Cell Receptor (TCR) play significant role in function of T cells and formation of the immunological synapse[46]. TCR pathway was reported to be significant in regulation of UC. Research has found that the increased levels of TCR core fucosylation are required for activating TCR signaling and inducing UC in mice, which might be a therapeutic strategy to block the process of UC formation[47]. By using transcriptional profiling of circulating T cells isolated from patients with UC, scientists considered that the prognosis in patients with UC could be predicted by gene expression profiling of Cluster of Differentiation 8 (CD8+) T cells[48]. As a pattern recognition receptors TLRs are crucial in immune response. TLRs playing a key role in identifying invading pathogens and upregulating signals related to inflammatory cytokines and costimulatory molecules[49]. An experimental study found that TLR4 deficiency aggravated intestinal injury in UC by down-regulating IL-6, CCL2 and CSF3[50]. NF-κB induces cytokine expression and neutrophil aggregation. NF-κB signaling pathway is a complex network which is involved in the processes of immune and inflammatory responses[51]. It is well accepted that activating NF-κB signaling pathway plays a key role in the development of UC and NF-κB regulates the expression of multiple pro-inflammation genes and maintains immune system homeostasis[52]. By suppressing NF-κB signaling pathway, the colonic glandular structure destruction and inflammatory cell infiltration of UC could be reduced, thus protecting epithelial barrier integrity[53].
In summary, the prediction results uncover that HLJDD has multiple ingredients, multiple targets and multiple approaches in treating UC, which enhance our understanding of the potential anti-inflammation mechanisms of HLJDD. However, this study also has some limitations on account of insufficient retrieval data and lacking in relevant experiments to validate the prediction results. Therefore, there is a lot of unfinished work needed for researchers to improve and accomplish to clarify the mechanism of HLJDD against UC.
From the network pharmacology prediction, we could conclude the active compounds, potential targets and possible mechanisms of HLJDD treating UC. 55 constituent compounds of HLJDD were summarized and filtered from TCMSP. 84 targets were identified as the common targets of UC and compounds. By network construction and topological calculation, 10 core active compounds and 10 core targets of HLJDD against UC were identified. Furthermore, possible MF, BP and pathways regulated by HLJDD against UC were systematically interpreted, which could provide a basis for its clinical application and further research.
Funding:
This study was supported by the Hebei University of Chinese Medicine 2021 Graduate Innovation Funding Project [Grant number: XCXZZBS2021004], S and T Program of Hebei [Grant numbers: 21377724D and 21377740D], Research Project of Hebei Provincial Administration of TCM [Grant number: 2022077), the Program of the National famous TCM inheritance Studio of the National Administration of TCM of China ((2018) No.119), and the Scientific Research Program of Hebei Administration of TCM (Grant number: 2022334).
Conflict of interests:
The authors declared no conflict of interests.
References
- Baumgart DC, Carding SR. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007;369(9573):1627-40.
- Cosnes J, Gower–Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011;140(6):1785-94.
[Crossref] [Google Scholar] [PubMed]
- Wei SC, Sollano J, Hui YT, Yu W, Santos Estrella PV, Llamado LJ, et al. Epidemiology, burden of disease and unmet needs in the treatment of ulcerative colitis in Asia. Expe Rev Gastroenterol Hepatol 2021;15(3):275-89.
[Crossref] [Google Scholar] [PubMed]
- Li X, Song P, Li J, Tao Y, Li G, Li X, et al. The disease burden and clinical characteristics of inflammatory bowel disease in the Chinese population: A systematic review and meta-analysis. Int J Environ Res Public Health 2017;14(3):238.
[Crossref] [Google Scholar] [PubMed]
- Kobayashi T, Siegmund B, Le Berre C. Ulcerative colitis. Nat Rev Dis Primers 2020;6(1):74.
- Hu HY. Evaluation of medication on ulcerative colitis and experiences of TCM in its treatment. Zhongguo Zhong Xi Yi Jie He Za Zhi 2008;28(9):780-2.
[Google Scholar] [PubMed]
- Qi Y, Zhang Q, Zhu H. Huang-LianJie-Du decoction: A review on phytochemical, pharmacological and pharmacokinetic investigations. Chin Med 2019;14(1):1-22.
- Yuan Z, Yang L, Zhang X, Ji P, Wei Y. Therapeutic effect of n-butanol fraction of Huang-lian-Jie-du decoction on ulcerative colitis and its regulation on intestinal flora in colitis mice. Biomed Pharmacother 2020;121:109638.
[Crossref] [Google Scholar] [PubMed]
- Cui XJ, Lu Z, Xiao SM, Fang XW, Wen YL, Xiong WN, et al. Anti-inflammatory effect, plasma effective components and therapeutic targets of Huanglian Jiedu decoction on ulcerative colitis mice. Zhongguo Zhong Yao Za Zhi 2021;46(1):206-13.
[Crossref] [Google Scholar] [PubMed]
- Hopkins AL. Network pharmacology. Nat Biotechnol 2007;25(10):1110-1.
[Crossref] [Google Scholar] [PubMed]
- Li J, Lu C, Jiang M, Niu X, Guo H, Li L, et al. Traditional Chinese medicine-based network pharmacology could lead to new multicompound drug discovery. Evid Based Complement Altern Med 2012;2012:149762.
[Crossref] [Google Scholar] [PubMed]
- Li S, Fan TP, Jia W, Lu A, Zhang W. Network pharmacology in traditional Chinese medicine. Evid Based Complement Altern Med 2014;2014.
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13.
[Crossref] [Google Scholar] [PubMed]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 1997;23(1-3):3-25.
[Crossref] [Google Scholar] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase. Nucl Acids Res 2018;46(5):2699.
- Sulakhe D, Taylor A, Balasubramanian S, Feng B, Xie B, Börnigen D, et al. Lynx web services for annotations and systems analysis of multi-gene disorders. Nucleic Acids Res 2014;42(W1):W473-7.
[Crossref] [Google Scholar] [PubMed]
- Fishilevich S, Zimmerman S, Kohn A, Iny Stein T, Olender T, Kolker E, et al. Genic insights from integrated human proteomics in GeneCards. Database 2016;2016.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Queralt-Rosinach N, Bravo A, Deu-Pons J, Bauer-Mehren A, Baron M, et al. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015;2015.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. 2017;45(D1):D362-8.
[Crossref] [Google Scholar] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011;27(3):431-2.
[Crossref] [Google Scholar] [PubMed]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization and integrated discovery. Genome Biol 2003;4(9):p3.
[Crossref] [Google Scholar] [PubMed]
- Parasuraman S. Protein data bank. J Pharmacol Pharmacother 2012;3(4):351.
[Google Scholar] [PubMed]
- Yuan Z, Yang L, Zhang X, Ji P, Hua Y, Wei Y. Mechanism of Huang-lian-Jie-du decoction and its effective fraction in alleviating acute ulcerative colitis in mice: Regulating arachidonic acid metabolism and glycerophospholipid metabolism. J Ethnopharmacol 2020;259:112872.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Yao J, Han C, Yang J, Chaudhry MT, Wang S, et al. Quercetin, inflammation and immunity. Nutrients 2016;8(3):167.
[Crossref] [Google Scholar] [PubMed]
- Dicarlo M, Teti G, Verna G, Liso M, Cavalcanti E, Sila A, et al. Quercetin exposure suppresses the inflammatory pathway in intestinal organoids from Winnie mice. Int J Mol Sci 2019;20(22):5771.
[Crossref] [Google Scholar] [PubMed]
- Guazelli CF, Fattori V, Colombo BB, Georgetti SR, Vicentini FT, Casagrande R, et al. Quercetin-loaded microcapsules ameliorate experimental colitis in mice by anti-inflammatory and antioxidant mechanisms. J Nat Prod 2013;76(2):200-8.
[Crossref] [Google Scholar] [PubMed]
- Devi KP, Malar DS, Nabavi SF, Sureda A, Xiao J, Nabavi SM, et al. Kaempferol and inflammation: From chemistry to medicine. Pharmacol Res 2015;99:1-0.
[Crossref] [Google Scholar] [PubMed]
- Park MY, JiGE, Sung MK. Dietary kaempferol suppresses inflammation of dextran sulfate sodium-induced colitis in mice. Dig Dis Sci 2012;57(2):355-63.
[Crossref] [Google Scholar] [PubMed]
- Sharifi Rad J, Herrera Bravo J, Salazar LA, Shaheen S, Abdulmajid Ayatollahi S, Kobarfard F, et al . The therapeutic potential of wogonin observed in preclinical studies. Evid Based Complement Alternat Med 2021;2021:9935451.
[Crossref] [Google Scholar] [PubMed]
- Wu Q, Xie S, Zhu Y, Chen J, Tian J, Xiong S, et al. Wogonin strengthens the therapeutic effects of mesenchymal stem cells in DSS induced colitis via promoting IL-10 production. Oxid Med Cell Longev 2021;2021:5527935.
- Li YY, Wang XJ, Su YL, Wang Q, Huang SW, Pan ZF, et al. Baicalein ameliorates ulcerative colitis by improving intestinal epithelial barrier via AhR/IL-22 pathway in ILC3s. Acta Pharmacol Sin 2022;43(6):1495-507.
[Crossref] [Google Scholar] [PubMed]
- Liu C, Li Y, Chen Y, Huang S, Wang X, Luo S, et al. Baicalein restores the balance of Th17/Treg cells via aryl hydrocarbon receptor to attenuate colitis. Mediat Inflamm 2020;2020:5918587.
[Crossref] [Google Scholar] [PubMed]
- Ren J, Yue B, Wang H, Zhang B, Luo X, Yu Z, et al. Acacetin ameliorates experimental colitis in mice via inhibiting macrophage inflammatory response and regulating the composition of gut microbiota. Front Physiol 2021;11:577237.
[Crossref] [Google Scholar] [PubMed]
- Carey R, Jurickova I, Ballard E, Bonkowski E, Han X, Xu H, et al. Activation of an IL-6: STAT3-dependent transcriptome in pediatric onset inflammatory bowel disease. Inflamm Bowel Dis 2008;14(4):446-57.
[Crossref] [Google Scholar] [PubMed]
- Nemetz A, Nosti-Escanilla MP, Molnár T, Köpe A, Kovács Á, Fehér J, et al. IL1B gene polymorphisms influence the course and severity of inflammatory bowel disease. Immunogenetics 1999;49(6):527-31.
[Crossref] [Google Scholar] [PubMed]
- Hirsch D, Gaiser T. Crohn’s disease-associated colorectal carcinogenesis: TP53 mutations and copy number gains of chromosome arm 5p as (early) markers of tumor progression. Pathol 2018;39(2):253-61.
- Cox DG, Crusius JB, Peeters PH, Bueno-de-Mesquita HB, Peña AS, Canzian F. Haplotype of prostaglandin synthase 2/cyclooxygenase 2 is involved in the susceptibility to inflammatory bowel disease. World J Gastroenterol 2005;11(38):6003.
[Crossref] [Google Scholar] [PubMed]
- Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: Redundancy and specificity. Oncogene 2001;20(19):2378-89.
[Crossref] [Google Scholar] [PubMed]
- Fisher RC, Bellamkonda K, Molina LA, Xiang S, Liska D, Sarvestani SK, et al. Disrupting inflammation-associated CXCL8-CXCR1 signaling inhibits tumorigenicity initiated by sporadic and colitis-colon cancer stem cells. Neoplasia 2019;21(3):269-81.
[Crossref] [Google Scholar] [PubMed]
- Venakteshaiah SU, Kumar KH. Inflammation and cancer. Endocr Metab Immune Disord Drug Targets 2021;21(2):193-4.
- Rhodes JM, Campbell BJ. Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol Med 2002;8(1):10-6.
[Crossref] [Google Scholar] [PubMed]
- Ekbom A, Helmick C, Zack M, Adami HO. Ulcerative colitis and colorectal cancer: A population-based study. N Engl J Med 1990;323(18):1228-33.
[Crossref] [Google Scholar] [PubMed]
- Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001;48(4):526-35.
[Crossref] [Google Scholar] [PubMed]
- Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol 2016;12(1):49-62.
[Crossref] [Google Scholar] [PubMed]
- Pugliese D, Felice C, Papa A, Gasbarrini A, Rapaccini GL, Guidi L, et al. Anti TNF-α therapy for ulcerative colitis: Current status and prospects for the future. Exp Rev Clin Immunol 2017;13(3):223-33.
[Crossref] [Google Scholar] [PubMed]
- Yokosuka T, Saito T. The immunological synapse, TCR microclusters and T cell activation. Curr Top Microbiol Immunol 2010;340:81-107.
[Crossref] [Google Scholar] [PubMed]
- Fujii H, Shinzaki S, Iijima H, Wakamatsu K, Iwamoto C, Sobajima T, et al. Core fucosylation on T cells, required for activation of T-cell receptor signaling and induction of colitis in mice, is increased in patients with inflammatory bowel disease. Gastroenterology 2016;150(7):1620-32.
[Crossref] [Google Scholar] [PubMed]
- Lee JC, Lyons PA, McKinney EF, Sowerby JM, Carr EJ, Bredin F, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Investig 2011;121(10):4170-9.
[Crossref] [Google Scholar] [PubMed]
- Kaisho T, Akira S. Toll-like receptors and their signaling mechanism in innate immunity. Acta Odontol Scand 2001;59(3):124-30.
- Shi YJ, Hu SJ, Zhao QQ, Liu XS, Liu C, Wang H. Toll-like receptor 4 (TLR4) deficiency aggravates dextran sulfate sodium (DSS) induced intestinal injury by down-regulating IL-6, CCL2 and CSF3. Ann Transl Med 2019;7(23).
[Crossref] [Google Scholar] [PubMed]
- Wang S, Liu Z, Wang L, Zhang X. NF-κB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol 2009;6(5):327-34.
[Crossref] [Google Scholar] [PubMed]
- Papadakis KA, Targan SR. Role of cytokines in the pathogenesis of inflammatory bowel disease. Ann Rev Med 2000;51(1):289-98.
[Crossref] [Google Scholar] [PubMed]
- Zeng L, Tan J, Xue M, Liu L, Wang M, Liang L, et al. An engineering probiotic producing defensin-5 ameliorating dextran sodium sulfate induced mice colitis via inhibiting NF-κB pathway. J Transl Med 2020;18:1-5.
 ): BP; (
): BP; ( ): CC and (
): CC and ( ): MF
): MF