- *Corresponding Author:
- Hui Mao
Jinhua Polytechnic
Jinhua321007, Zhejiang, China
E-mail: maohui1988@126.com
| Date of Received | 28 February 2020 |
| Date of Revision | 29 March 2021 |
| Date of Acceptance | 08 February 2022 |
| Indian J Pharm Sci 2022;84(1):173-181 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Molecular docking technology was employed to predict and exploit potential main protein inhibitors of novel coronavirus ribonucleic acid dependent ribonucleic acid polymerase by virtual screening of twenty hundred thousand natural molecules in ZINC database. By targeting main protease of novel coronavirus by Schrodinger Maestro software and molecular dynamic simulation, the affinity and stability of the complex formed between the compound and the main protease of novel coronavirus were carefully analyzed. Base on high-throughput virtual screening, twelve compounds with higher molecular docking score were selected from twenty hundred thousand compounds database, compound ZINC000096222420 has the highest docking score of -8.693. The results from molecular dynamic simulation and binding free energy calculation reveal that the structure of the complex is highly stable, which has high potential to accelerate the development of anti-severe acute respiratory syndrome coronavirus 2 drugs.
Keywords
Novel coronavirus, virtual screening, molecular dynamic simulation, small molecular compounds, main protease protein
Severe Acute Respiratory Syndrome Corona Virus (SARS-CoV) is a single strand plus Ribonucleic acid (RNA) virus that is infectious to vertebrates such as humans and livestock[1]. It was firstly discovered from an unusual inflammation of acute respiratory infections in 2003, which was subsequently officially named by the World Health Organization (WHO) as Severe Acute Respiratory Syndrome (SARS). The epidemic caused about 8000 infections worldwide and led to a mortality rate of up to about 10 %[2]. The outbreak of new Coronavirus Disease (COVID-19) in Wuhan, China, in December 2019 was also caused by another new coronavirus SARS-CoV-2, which has high homology with SARS-CoV. As of February 26, 2021, there are more than 113.85 million confirmed cases of new coronary pneumonia in the world, with more than 2.5 million deaths, which poses a great threat to human health[3-5]. The main transmission route of SARS virus is respiratory droplets and contact infection with fast transmission speed and high infection rate. Main symptoms of COVID-19 include fever, sore muscles, cough, sore throat and respiratory distress syndrome for severe cases. Unlike previous outbreaks, the increase in the proportion of patients without any respiratory symptoms also adds difficulty to the prevention and control of the epidemic. During the research process for seeking anti-SARS-CoV-2 drugs, the emergence of such drugs as chloroquine, hydroxychloroquine may reduce the mortality of the new coronavirus[6]. However, by far, there are no effective drugs targeting for SARS viruses on the market. The spread and infection of virus has seriously affected human health and economic development, so the development of relevant vaccines and therapeutic drugs is imminent[7].
With no doubt, vaccines are one of the effective treatments to prevent and control infectious diseases, but there are many difficulties in the development of efficient coronavirus vaccine. SARS-CoV-2 is highly glycosylated spherical particles with 66 to 87 glycosylation sites in the surface protein shell. Glycosylation protects specific epitopes from antibody neutralization and helps the virus escape immunity[8,9]. Exploiting efficient and inexpensive antiviral vaccines remains a great challenge. By comparison, small molecular compounds can more easily avoid polysaccharide coverage and thus provide effective treatments[10].
The Main protease (Mpro) of the novel coronavirus (SARS-CoV-2) is a class of cysteine hydrolases that cleaves protein precursors for viral genome translation, accomplishing normal transcription and replication[11-13]. The protein has highly conserved three-dimensional structure in a variety of corona viruses. Its conserved nature and lack of human homologs provide a promising target for the development of broad-spectrum anti-coronavirus therapeutic agents[14]. Hotada et al.[15] reported the interaction analysis of Mpro and its peptide like inhibitor N3 complex based on fragment molecular orbital, and revealed that His41, His163, His164 and Glu166 are the most important amino acid residues in the interaction between Mpro and inhibitors, which can form hydrogen bonds with inhibitors. Yang et al.[16] constructed a homology model for SARS-CoV-2 Mpro and found that N3 could fit inside the substrate-binding pocket as a time-dependent irreversible inhibitor of this enzyme. Recently, Hotada et al.[17] again calculated interactions within the complex formed between the main protease of the new coronavirus and the inhibitor N3 by employing the combination of classical Molecular Dynamics (MD) simulation and ab initio Fragment Molecular Orbital (FMO), and found that His163 is the outstanding residue with the largest stabilization in both “static” and “dynamic” evaluations, with other significantly stable residues including Met165, Leu167 and Gln189. Tien et al.[18] disclosed an important ligand binding mechanism in Mpro, implying that ligand binding stability in Mpro pockets can be markedly enhanced if the hydrophobic group of the ligand occupies its so-called "anchor point". Currently, the mortality rate of COVID-19 is still high, so the increasing relevant research needs to br carried out to exploit potential active inhibitors.
Materials and Methods
Virtual screening software and materials:
Molecular docking in this research was performed by employing Maestro software (from Schrodinger 2020- 2). The crystal structure of protein was obtained from the Protein Data Bank (PDB) database (https://www.rcsb.org) and the structure of the compound comes from the natural small molecular compounds of the ZINC compound library with a total number of 200 thousand.
Experimental methods:
Treatment of SARS-CoV-2-Mpro protein crystal structure: The three-dimensional crystal structure of the Mpro protein was gained and downloaded from the PDB database (PDB ID: 6LU7) with a resolution of 2.16 Å. The “Protein Preparation Wizard” of Maestro software was used to make modification of the chemical bonds, hydrogenation, treatment of metal ions, repair of missing atoms and amino acid residues, deletion of redundant molecules, etc. After energy optimization under the condition of Optimized Potentials for Liquid Simulations (OPLS) force field, the protein was finally selected to be the receptor of molecular docking.
Drug binding sites:
6LU7 is complex crystal structure of Mpro and compound N3. The docking target is based on the active pocket defined by the original ligand inhibitor N3 in the crystal complex and the docking grid is produced by receptor grid generation in Maestro software.
Treatment of compound structure:
A total number of 200 000 small molecules of natural compounds were provided by the ZINC database. A “Ligprep” module is applied to optimize the compound molecules (Parameter setting: pH=7, Epik option was used to calculate the protonation state of the molecule, generated at most 32 per ligand).
Molecular docking:
6LU7 was used as rigid receptors and optimized compound molecules were as flexible ligands for semi-flexible docking. “Ligand Docking” modules are applied for high throughput virtual screening. The docking score was employed as screening condition and the complex of SARS-Cov-2-Mpro protein and candidate drug was constructed for MD simulation.
MD simulation:
The MD simulation is carried out by GPU accelerated pmemd.cuda modules in the AMBER software package. Protonation state of titratable protein residues can be predicted by H++ at pH 7.0. The His41 and His80 are Nδ protonated, while the His64, His163, His164, His172 and His246 are Nε protonated. The protein was hydrogenated using the "reduce" program of AmberTools18, which was then visually examined by a PyMOL procedure. The selected General Amber Force Field (gaff)2 force field was added to the small molecular substrate. The electrostatic potential of the substrate was calculated at the HF/6-31G* theoretical level and the Restrained Electrostatic Potential (RESP) charge was calculated using the Multiwfn3.7. The atomic types and parameters of all substrates can be obtained by AnteChamber PYthon Parser interfacE (ACPYPE). Subsequently, the protein-small molecule complex structure was immersed in Transferable Intermolecular Potential with 3 Points (TIP3P) water box so that the edge distance from any protein atom was at least 12 Å and the added countervailing ions (Na+ and Cl-) neutralized the charge. The protein and ion parameters are described by the ff14SB force field. The cutoff values for calculating Lennard-Jones and coulomb interactions, calculating long-range electrostatic interactions using Particle Mesh Ewald (PME) methods and constraining hydrogen-containing bonds using SHAKE algorithms are all set to 10 Å.
In order to eliminate bad contact, three stages of energy minimization are performed to optimize the geometry. Firstly, the whole protein, cofactor and substrate were constrained by a force constant of 100 kcal/mol·Å-2 for 5000 cycles (2500 steepest descent method, followed by the balance in conjugate gradient). Then, the skeleton atoms of protein were constrained by a force constant of 10 kcal/mol·Å-2, and 5000 steps of energy minimization (2500 steepest descent and 2500 steps conjugate gradient method) were performed. Finally, the whole system performs 10 000 steps of energy minimization without any constraints (5000 steepest descent and 5000 steps conjugate gradient method). After optimizing the structure, 100 ps was heated to 300 K at a constant volume. Langevin thermostat was utilized to adjust the temperature. The coupling time constant was 1.0 ps, and the enzyme, cofactor and substrate were limited by 10 kcal/mol·Å-2 position constraint. 5 kcal/mol·Å-2 position constraint was used for density balance at constant pressure for 500 ps, followed by an unconstrained 1 ns NPT simulation, where both temperature coupling and pressure relaxation times are set to 5 ps. MD simulations of 50 ns were performed in the NPT ensemble after 300 K equilibria, where the Langevin thermostat was employed to maintain the temperature and the pressure was controlled at 1.0 atm employing Monte Carlo barostat. The simulation time step is 2 fs, and the trajectory is saved every 10 ps. The MD trajectory is analyzed and post-processed using CPPTRAJ program, and visual inspection is carried out using visual MD.
Binding free energy calculation:
Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) or Molecular Mechanics Poisson- Boltzmann Surface Area (MM-PBSA) method was utilized to calculate and decompose the MD simulation data (300 frames) at 20-50 ns in the equilibrium state of the system are calculated. The decomposition process consists of four energy types: ΔGbind=+ΔEele+ΔGpol+ΔGnopol. Among them, ΔEvdw represents nonbonding van der waals interactions, ΔEele represents electrostatic interactions, and ΔGpol and ΔGnopol represent polar and nonpolar interactions, respectively, which constitute the solvation free energy. The igb is set to 5 in the input file, and other parameters are default. MM-GBSA calculation is carried out using the MMPBSA.py program in the AmberTools 18.
Results and Discussion
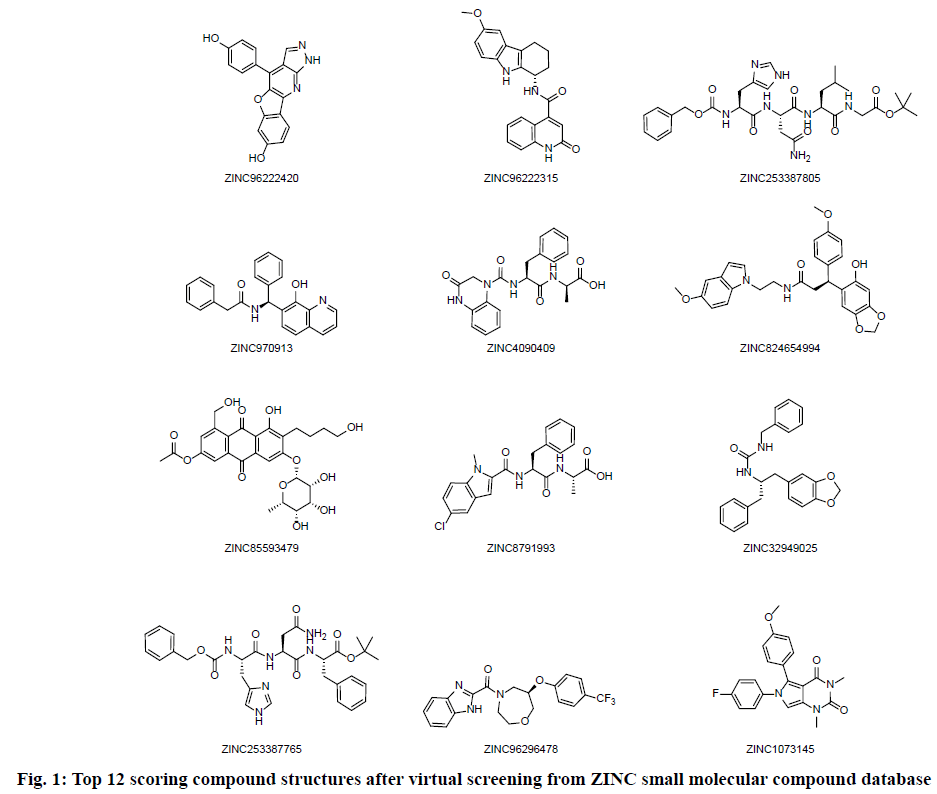
High throughput screening was carried out from 200 thousands ZINC small molecular compound database and twelve compounds with highest molecular docking score were selected. The screening results are shown in Table 1 and the structure of the compounds is shown in fig. 1. The four compounds ZINC96222420, ZINC96222315, ZINC253387805 and ZINC4090409 with better docking score were selected for further analysis.
| Number | Name | MW | Log P | Donor HB | Accept HB | Docking score |
|---|---|---|---|---|---|---|
| 1 | ZINC96222420 | 317.30 | 3.936 | 3 | 5 | -8.963 |
| 2 | ZINC96222315 | 387.44 | 3.825 | 3 | 3 | -8.807 |
| 3 | ZINC253387805 | 629.72 | 0.596 | 6 | 9 | -8.756 |
| 4 | ZINC970913 | 368.44 | 4.389 | 2 | 3 | -8.713 |
| 5 | ZINC4090409 | 410.43 | 1.355 | 3 | 5 | -8.676 |
| 6 | ZINC824654994 | 488.54 | 4.431 | 2 | 6 | -8.663 |
| 7 | ZINC85593479 | 546.53 | 0.106 | 6 | 6 | -8.662 |
| 8 | ZINC8791993 | 426.88 | 2.762 | 2 | 5 | -8.614 |
| 9 | ZINC32949025 | 388.47 | 4.069 | 2 | 3 | -8.613 |
| 10 | ZINC253387765 | 607.69 | 1.677 | 5 | 8 | -8.457 |
| 11 | ZINC96296478 | 405.38 | 3.502 | 1 | 4 | -8.403 |
| 12 | ZINC1073145 | 379.39 | 2.843 | 0 | 6 | -8.315 |
Table 1: Virtual Screening Results of Drug Candidates
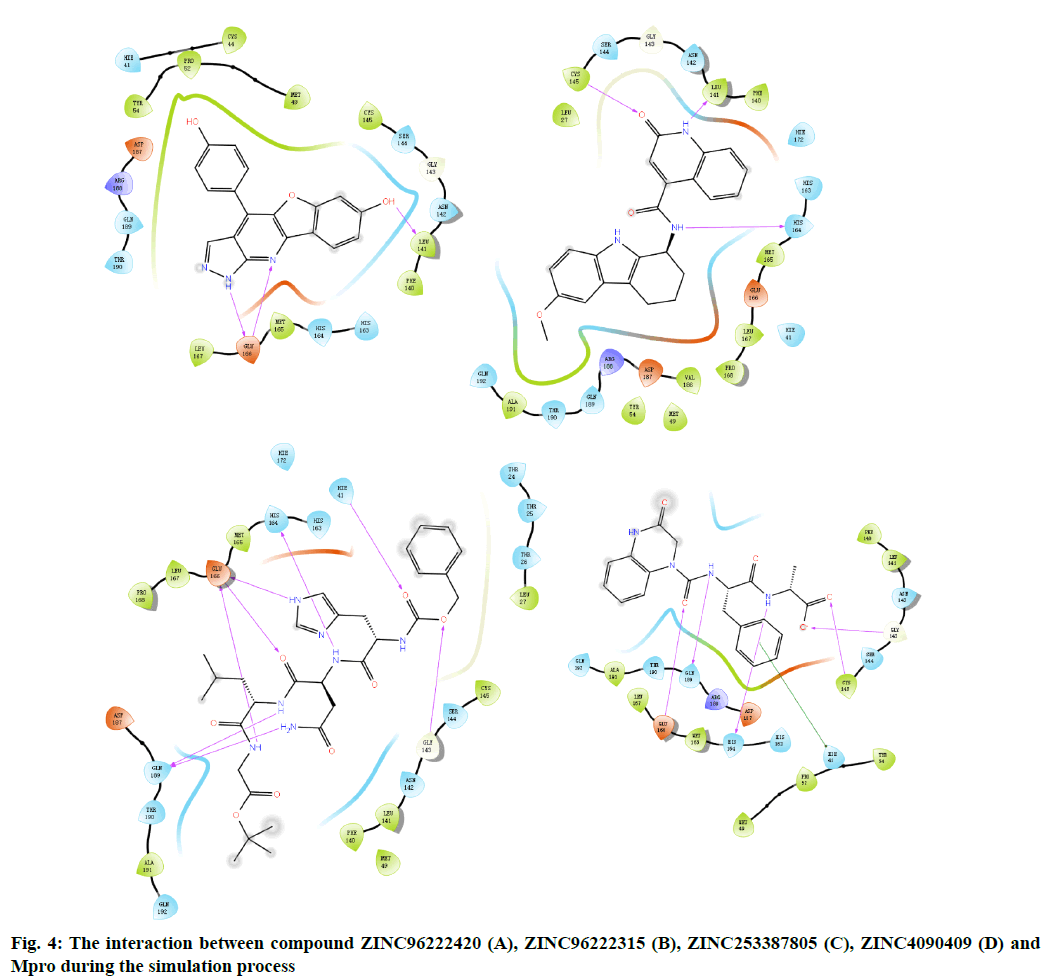
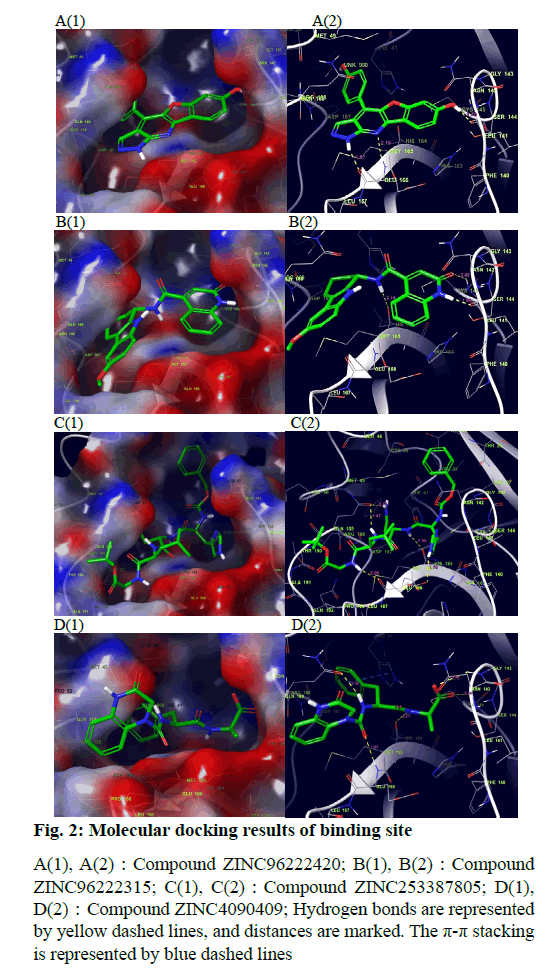
The docking result of small molecule ZINC96222420 to target protein molecule is shown in fig. 2A. The docking score of the compound was -8.963 kcal/mol. The phenol fragments of small molecules extend into the active cavity composed of Met49, Pro52, Asp187 and Arg188. The nitrogen atoms on the six-member nitrogen heterocycle and the imino hydrogen on the five-member nitrogen heterocycle can form hydrogen bonds with the Glu166 with bond length of 2.19 Å and 1.87 Å respectively. The hydrogen of the hydroxyl group on the benzene ring adjacent to the oxygencontaining five-member ring can form a hydrogen bond with the Leu141 and the a bond length is 1.70 Å.
Fig. 2: Molecular docking results of binding site
A(1), A(2):Compound ZINC96222420; B(1), B(2):Compound
ZINC96222315; C(1), C(2):Compound ZINC253387805; D(1),
D(2):Compound ZINC4090409; Hydrogen bonds are represented
by yellow dashed lines, and distances are marked. The p-p stacking
is represented by blue dashed lines
The docking result of small molecule ZINC96222315 to target protein molecule is shown in fig. 2B. The docking score of the compound was -8.807 kcal/ mol. The hydrogen atom on the peptide bond forms a hydrogen bond with the His164 with bond length of 2.19 Å. The hydrogen bond forms between the imino hydrogen atom of six-member nitrogen heterocycle and leu141 with bond length of 1.94 Å. The carbonyl oxygen atom on the six-member nitrogen heterocycle forms hydrogen bond with cys145 with a bond length of 2.02 Å.
The docking result of small molecule ZINC253387805 to target protein molecule is shown in fig. 2C. The docking score of the compound was -8.756 kcal/mol. The amino hydrogen atom on glycine, oxygen atom on asparagine main chain and imino hydrogen atom on imidazole group of histidine side chain all form hydrogen bonds with Glu166 with bond length of 2.09 Å, 2.03 Å and 2.75 Å respectively. The amino hydrogen atom on leucine and the amino hydrogen atom on the side chain of asparagine form hydrogen bonds with Gln189 with bond length of 2.47 Å and 2.41 Å, respectively. The imino hydrogen atom on asparagine forms a hydrogen bond with His164 with a bond length of 2.30 Å. The oxygen atom adjacent to the benzene ring forms a hydrogen bond with Gly143 with a bond length of 2.64 Å and the oxygen atom of peptide bond adjacent to the benzene ring forms hydrogen bond with Hie41 and the bond length is 2.49 Å.
The docking result of small molecule ZINC4090409 to target protein molecule is shown in fig. 2D. The docking score of the compound was -8.676 kcal/mol. The oxygen atom of peptide bond adjacent to six-member nitrogen heterocycle forms hydrogen bond with Glu 166 with bond length of 1.93 Å. The amino hydrogen atom on phenylalanine forms a hydrogen bond with Gln189 with bond length of 2.20 Å. The amino hydrogen atom on alanine forms a hydrogen bond with the His164 with bond length of 2.27 Å, while the oxygen atom on alanine forms a hydrogen bond with the Cys145, and the bond length is 2.24 Å. The oxygen atom at the end of the small molecule forms a hydrogen bond with the Gly143 with a bond length of 2.70 Å. The benzene ring on phenylalanine forms stable π-π stacking interaction with Hie41.
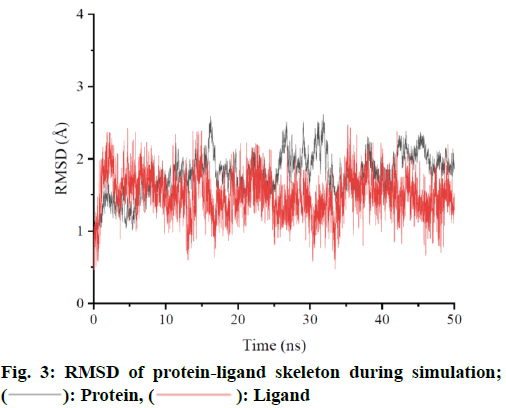
MD simulation was performed for ZINC962222420, which ranked the highest score among the four compounds. In order to ensure the dynamic stability and sampling rationality, Root Mean Square Deviation (RMSD) values of protein skeleton atoms were calculated based on crystal structure (fig. 3). If the oscillation amplitude of the overall RMSD value is less than 1.0 Å, the system is in equilibrium. The RMSD diagram shows that the complex reaches equilibrium at about 5 ns and is stable at about 1.8 Å. The RMSD was low during the simulation process, indicating that the complex formed was stable. Based on the RMSD of the protein skeleton, it is found that the RMSD of the ligand is also stable and has little fluctuation in the whole simulation process. The average conformation (fig. 4) shows that the binding mode of ligand molecules to receptors is consistent with the initial state during the simulation.
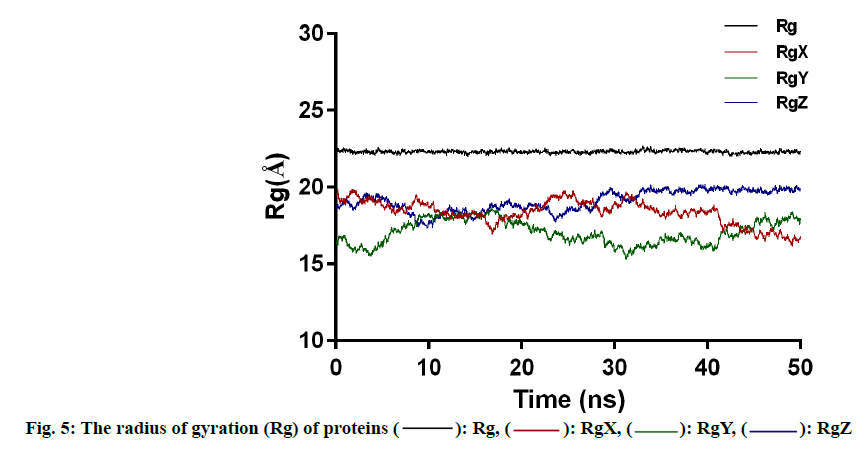
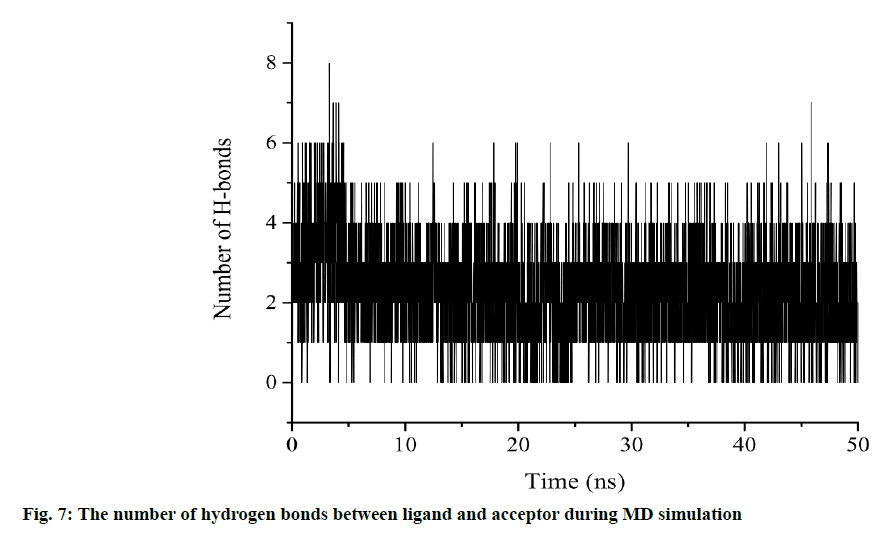
Another characterization of the stability of MD system is the Radius of Gyration (Rg) of proteins, whose fluctuation range reflects the degree of protein density. The larger the value, the more relaxed the protein is. In this study, the total Rg values and the Rg values of each dimension are shown in fig. 5. Among them, the total Rg remained basically unchanged, indicating that the protein remained stable and dense during the whole simulation process. The final conformation of the complex overlaps with the initial conformation (fig. 6), indicating that the conformational deviation of both protein and inhibitor is small. These results show that the complex system remains stable and can be further analyzed. The heat diagram of intermolecular hydrogen bond between receptor and ligand is shown in fig. 7. During the kinetic simulation, the system maintains more than two intermolecular hydrogen bonds on average, which is consistent with the docking results.
The contribution of the MM-GBSA binding free energy of the ZINC96222420 to the target protein is listed in Table 2. The total binding energy of the complex is -30.2352 kcal/mol and the low binding energy confirms the stability of the complex. The net gas phase energy is -59.185 kcal/mol. For this energy, van der Waals was the main contributor (-35.5349 kcal/mol). Electrostatic energy plays a relatively small role in binding (-23.6636 kcal/mol). The total solvation energy of MM-GBSA is 28.9633 kcal/mol. In addition, the binding energy was decomposed to specific amino acids, focusing on the contribution of amino acids interacting with inhibitors in the docking model. The binding free energies of hotspot receptor residues interacting with inhibitors during MD simulations (The contribution of binding energy is more than 0.5 kcal/mol) are shown below. They are highly consistent with the docking model, indicating that the model has good reliability.
| Complex | |||
|---|---|---|---|
| Energy | Component | Average Std. Dev. | Std. Err. of Mean |
| VDWAALS | -2433.8426 | 19.2168 | 1.9121 |
| EEL | -21335.4238 | 87.4131 | 8.6979 |
| EGB | -2966.4105 | 68.507 | 6.8167 |
| ESURF | 102.8384 | 1.6621 | 0.1654 |
| G gas | -23769.2664 | 87.4035 | 8.697 |
| G solv | -2863.5722 | 67.5442 | 6.7209 |
| TOTAL | -26632.8386 | 58.6805 | 5.8389 |
| Receptor | |||
| VDWAALS | -2394.1488 | 18.8935 | 1.88 |
| EEL | -21404.6864 | 87.8289 | 8.7393 |
| EGB | -2973.6515 | 68.5732 | 6.8233 |
| ESURF | 104.5158 | 1.6504 | 0.1642 |
| G gas | -23798.8351 | 87.5519 | 8.7117 |
| G solv | -2869.1357 | 67.6234 | 6.7288 |
| TOTAL | -26667.9709 | 57.9676 | 5.768 |
| Ligand | |||
| VDWAALS | -4.159 | 0.294 | 0.0292 |
| EEL | 92.9262 | 1.511 | 0.1503 |
| EGB | -26.1268 | 0.4179 | 0.0416 |
| ESURF | 2.7271 | 0.013 | 0.0013 |
| G gas | 88.7672 | 1.4891 | 0.1482 |
| G solv | -23.3997 | 0.4134 | 0.0411 |
| TOTAL | 65.3675 | 1.6057 | 0.1598 |
| Differences (Complex-Receptor-Ligand) | |||
| VDWAALS | -35.5349 | 2.6932 | 0.268 |
| EEL | -23.6636 | 5.0916 | 0.5066 |
| EGB | 33.3678 | 2.1219 | 0.2111 |
| ESURF | -4.4045 | 0.1796 | 0.0179 |
| DELTA G gas | -59.1985 | 4.4582 | 0.4436 |
| DELTA G solv | 28.9633 | 2.0683 | 0.2058 |
| DELTA TOTAL | -30.2352 | 3.4045 | 0.3388 |
Table 2: Binding Free Energy Calculation Based on MM-GBSA Method (Kcal/Mol)
Met165 (-2.5023 kcal/mol), Glu166 (-2.4046 kcal/mol), Asp187 (-1.9558 kcal/mol), Met49 (-1.2253 kcal/mol), Gln192 (-0.9701 kcal/mol), Arg188 (-0.9678 kcal/mol), Gln189 (-0.7984 kcal/mol), His41(-0.7678 kcal/mol), Cys145 (-0.7175 kcal/mol), Ser144 (-0.6901 kcal/mol), Asn142 (-0.6760 kcal/mol).
Mpro protease is the main functional protein of coronavirus such as SARS-CoV and SARS-CoV-2. In this study, the Mpro inhibitors of novel coronavirus were targeted for virtually screening from 200 thousand natural compounds in the ZINC15 database using molecular docking and MD methods. After analysis, the amino acid residue Glu166 plays an important role in the binding of ligands to SARS-CoV-2 Mpro, which is consistent with recent results. This may be the goal of optimizing the structure of compounds in the future[15,19]. The MD simulation of 50 ns aqueous solution shows that the binding conformation of ZINC96222420 compound and SARS-CoV-2 main protease complex is highly stable. The binding free energy calculated by MM-GBSA method also confirmed the stability of ZINC962222420 Mpro complex. Based on these results, this compound has high potential to accelerate the development of Anti-SARS-CoV-2 drugs. The enzyme kinetics experiments of related compounds with top docking scores will be carried on for further research aimed at predicting new antiviral drugs for the treatment of COVID-19.
Acknowledgments:
This project was supported by Key Research Projects on Emergency Prevention and Control of New Coronavirus Infected Pneumonia from Jinhua Science and Technology Bureau(2020XG-11). We would like to thank Dr. Xiang Fei of Gachon University for his great support in computer docking (Software: Maestro Software (from Schrodinger 2020-2)).
Author contributions:
Hui Mao and Chunxia Chu contributed equally to this work.
References
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17(3):181-92.
[Crossref] [Google Scholar] [PubMed]
- Severe Acute Respiratory Syndrome (SARS). World Health Organization; 2022.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579(7798):265-9.
[Crossref] [Google Scholar] [PubMed]
- COVID-19 Case Tracker. Johns Hopkins University and Medicine; 2021.
- Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci Chin Life Sci 2020;63(3):457-60.
[Crossref] [Google Scholar] [PubMed]
- Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents 2020;105932(10.1016).
[Crossref] [Google Scholar] [PubMed]
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 2020;19(3):149-50.
[Crossref] [Google Scholar] [PubMed]
- Watanabe Y, Berndsen ZT, Raghwani J, Seabright GE, Allen JD, Pybus OG, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun 2020;11(1):2688.
[Crossref] [Google Scholar] [PubMed]
- Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 2007;15(5):211-8.
[Crossref] [Google Scholar] [PubMed]
- Casalino L, Gaieb Z, Dommer AC, Harbison AM, Fogarty CA, Barros EP, et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cen Sci 2020;6:1722-34.
[Crossref] [Google Scholar] [PubMed]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020;582(7811):289-93.
[Crossref] [Google Scholar] [PubMed]
- Gimeno A, Mestres-Truyol J, Ojeda-Montes MJ, Macip G, Saldivar-Espinoza B, Cereto-Massagué A, et al. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int J Mol Sci 2020;21(11):3793.
[Crossref] [Google Scholar] [PubMed]
- Kalhotra P, Chittepu VC, Osorio-Revilla G, Gallardo-Velazquez T. Field-Template, QSAR, Ensemble Molecular Docking, and 3D-RISM Solvation Studies Expose Potential of FDA-Approved Marine Drugs as SARS-CoVID-2 Main Protease Inhibitors. Molecules 2021;26(4):936.
[Crossref] [Google Scholar] [PubMed]
- Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chem Biochem 2020;21(5):730-8.
[Crossref] [Google Scholar] [PubMed]
- Hatada R, Okuwaki K, Mochizuki Y, Handa Y, Fukuzawa K, Komeiji Y, et al. Fragment molecular orbital based interaction analyses on COVID-19 main protease- inhibitor N3 complex (PDB ID: 6LU7). J Chem Inf Model 2020;60(7):3593-602.
[Crossref] [Google Scholar] [PubMed]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020;582(7811):289-93.
[Crossref] [Google Scholar] [PubMed]
- Hatada R, Okuwaki K, Akisawa K, Mochizuki Y, Handa Y, Fukuzawa K, et al. Statistical interaction analyses between SARS-CoV-2 main protease and inhibitor N3 by combining molecular dynamics simulation and fragment molecular orbital calculation. Appl Phys Express 2021;14(2):027003.
- Huynh T, Wang H, Luan B. In silico exploration of the molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. J Phys Chem Lett 2020;11(11):4413-20.
[Crossref] [Google Scholar] [PubMed]
- Ngo ST, Quynh Anh Pham N, Thi Le L, Pham DH, Vu VV. Computational determination of potential inhibitors of SARS-CoV-2 main protease. J Chem Inf Model 2020;60(12):5771-80.
[Crossref] [Google Scholar] [PubMed]
 Ligand
Ligand