- *Corresponding Author:
- C. Fang
Department of Anesthesiology,
Third Xiangya Hospital of Central South University,
Changsha,
Hunan 410013,
China
E-mail: fangchao412@yeah.net
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “19-26” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cervical cancer has become a most lethal and extensive gynecological malignancy, mainly because of the complete absence of symptoms in the early stage of the disease and the lack of early diagnostic markers. Several studies have found that deoxyribonucleic acid methylation plays an important role in the development of cervical cancer without altering the sequence of genomic deoxyribonucleic acid. Zinc finger protein 582 is on chromosome 19 and contains a Kruppel-associated box-AB region and 9 zinc finger motifs. Detection of the zinc finger protein 582 methylation in cervical precancerous lesions is helpful for the early diagnosis of cervical cancer. The level of chemotherapy increased with the increase of lesion grade and it was hypermethylated in cervical cancer tissues. It is hypothesized that the role of zinc finger protein 582 is like that of the tumor suppressor gene and the tumor gene expression decreases with an increased zinc finger protein 582 methylation level. This study aimed to describe the research progress of the zinc finger protein 582 methylation, detection in cervical cancer.
Keywords
Deoxyribonucleic acid methylation, zinc finger protein 582, epigenetics, tumor suppressor gene, cervical cancer, biomarker
Regarding cervical cancer, global cancer data shows that there are 604 127 new cases and 341 831 deaths in 2020[1]. Human Papillomavirus (HPV) is almost necessary (but not sufficient) for cervical cancer throughout the entire cancer process. Its infection shows a primary risk factor that can be attributed to cervical cancer[2]. According to the latest report, benefit from the prevention of cancer screening and the spread of vaccines, in developed countries, the incidence rate and mortality rate of cervical cancer have declined. However, cervical cancer is still a great threat to women in developing countries[3]. Some studies have found that Deoxyribonucleic Acid (DNA) methylation plays an important role in the occurrence and development of cervical cancer[4-6]. Studies have documented the promise of Zinc Finger protein 582 (ZNF582) methylation in the detection of cervical precancerous lesions and methylation frequently silenced ZNF582 in cervical cancers[7,8]. Our previous studies showed that the ZNF582 methylation test improves cervical cancer screening efficacy[9,10]. We summarized the function of DNA methylation in cancer and highlighted whether methylation of ZNF582 contributes to the development of cervical cancer and early diagnosis in cervical cancer.
Epigenetics and Methylation
As Conrad Waddington defined, epigenetics is a branch of biology, which studies the interaction between genes and their phenotypic products[11], and DNA methylation is a most manifestation. DNA methylation makes up the interface that compromises a viral genome, including the environment. This leaves the genetic records of this interaction, which is an ideal biomarker for the detection of cancer[12]. In tumor cells, DNA hypermethylation in promoter regions can lead to the inactivation of tumor suppressor genes, whereas hypomethylation at genome- wide levels leads to increased genomic instability and cell transformation[13]. Based on the theory above, the researchers found gene methylation plays a role in the development and spontaneous regression of several tumors[14,15]. In mammals, DNA methylation is almost entirely present in Cytosines followed by Guanine residues (CpG) dinucleotides and 60 %~80 % of CpG in the entire genome of somatic cells is methylated, it is regarded to occur at specific positions[16]. With the deepening of methylation research, many hypomethylated genomic regions have recently been detected in the genomic region using genomic microarray. In tumor development, hypomethylation of genomic DNA increased with the development of a benign proliferation of cells to invasive cancer[17].
ZNF family and ZNF582:
ZNFs are a large and diverse family of proteins. ZNFs are often defined as proteins whose functional regions require at least one zinc ion coordination. Zinc ions are used to stabilize the integration of the proteins themselves and not involved in binding to the target gene[18]. Cys2 His2 is the first domain found in the ZNF family, comprising 30 amino acids and nine repeats, each domain folded to form a left-handed ββα secondary structure, two halves on a β-sheet hairpin. The zinc ion is coordinated between cysteine and the two histidines in the α-helix[19].
ZNF582, a member of the ZNF family, is on chromosome 19 and encodes a nuclear protein of ZNF582 containing a Kruppel-Associated Box (KRAB) domain and nine zinc finger motifs. ZNF582 is most expressed in thyroid tissue and testis tissue.
The ZNF582 protein is predicted to be an intracellular protein transcription factor with five transcripts. The epidermoid carcinoma cell line (A-431) and the human bone osteosarcoma epithelial cells (U2-OS) cell line were detected by immunofluorescence, and the ZNF582 protein was found to be in the nucleoplasm and cytoplasm[20].
The researchers also revealed that ZNF582 gene may be involved in DNA damage response, proliferation, cell cycle regulation and tumor transformation[21,22]. The other study also found that ZNF582 gene is silenced in invasive cervical cancer, but it is still expressed in precancerous lesions[8].
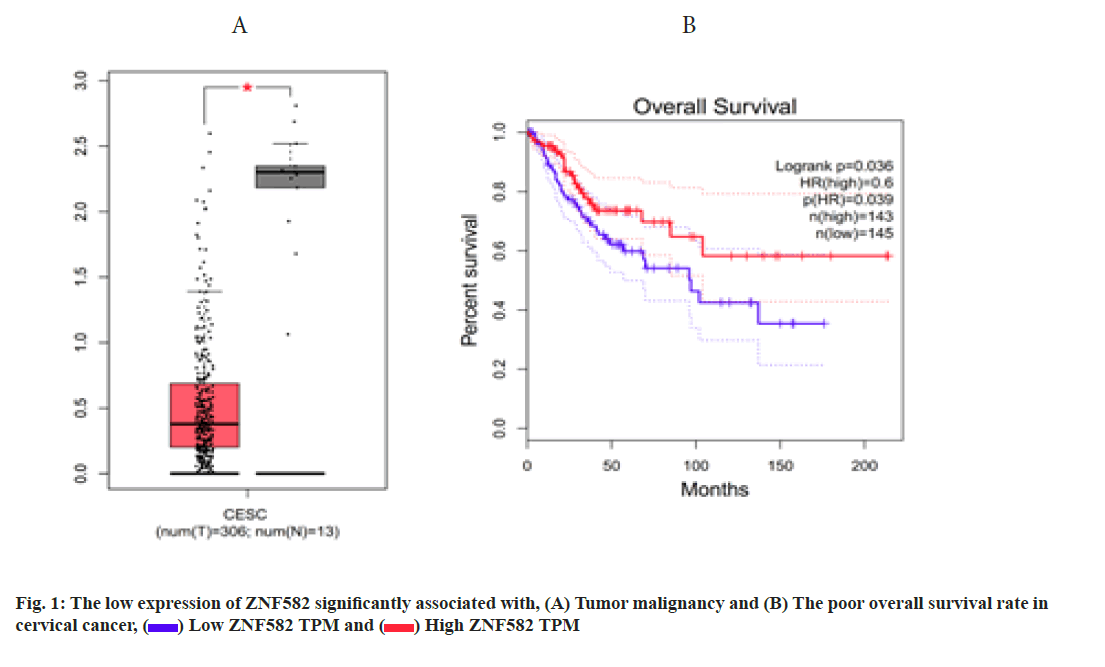
Bioinformatics analysis using cervical cancer case data in The Cancer Genome Atlas (TCGA) database (http:// gepia.cancer-pku.cn/detail.php?gene=ZNF582) found that the expression level of ZNF582 in cervical cancer tissues was significantly lower than that in normal tissues (p<0.05, fig. 1A) and the overall survival rate of patients with high ZNF582 expression was significantly better than that of patients with low expression (p<0.05, fig. 1B).
The Role of Gene Methylation in the Development of Cervical Cancer
Epigenetic silencing was found to be involved in cell cycle regulation, apoptosis, DNA repair, cell development and differentiation, hormone response, and signal transduction in clinical specimens representing different stages of cervical precancerous lesions and established cervical cancer cell lines[12,23]. Promoter- related sequence methylation characterized the change of tumor methylation pattern.
PR Domain zinc finger protein 14 (PRDM14) matters in invasive cervical development caused by high-risk HPV infection and HPV-induced cervix cancer and methylation-mediated PRDM14 gene silencing provide a new therapeutic target for it[24]. In vitro studies have found that cisplatin is like the demethylating agent 5-azacytidine and exerts a growth inhibitory effect on tumor cells by removing the methylation of Estrogen Receptor 1 (ESR1), Breast Cancer Susceptibility Protein-1 (BRCA1), RAS Association Domain Family Protein 1 Isoform A (RASSF1A), MutL Homolog 1 (MLH1), Myogenic Differentiation 1 (MYOD1), human Telomerase Reverse Transcriptase (hTERT) and Death-Associated Protein Kinase 1 (DAPK1)[25,26]. In addition, methylation modifications affect the sensitivity to chemotherapy and radiation therapy. Studies on a new Human cell line (SiHa) cells are more sensitive to radiation when they are treated with the combination of hydralazine, a DNA methylation inhibitor and valproic acid, a histone deacetylase inhibitor[27].
Although HPV infection is closely related to the occurrence of cervical cancer, only a few people infected with HPV progress to cervical cancer[28,29]. The persistent HPV infection does not transform and immortalize cervical epithelial cells. The genetic and epigenetic transformation disrupted cell cycle control, which is needed in the acquirement of the immortal phenotype and progress to the invasive phenotype. The transcription of the early HPV genomic region was regulated by the upstream of the Long Control Region (LCR), as the instability of the genome was produced by somatic mutations and gene recombination in the host genomic region[30]. Transcriptionally active HPV DNA copies are limited by epigenetic repression after the host cell has been immortalized, which only allows the minimal expression of the maintenance of immortalized phenotype necessary for viral oncoproteins[12]. Epigenetics not only regulate viral expression, it also plays a role in the progression of cervical cancer, so detecting changes in methylation has been away for tumor diagnosis, detecting of tumor progression and recurrence and may bring an opportunity for therapeutic value.
The recombination of HPV and chromosomal DNA was common in the development of cervical cancer and may be necessary for tumor progression. Kim et al. found that the activity of the viral promoter was affected by the methylation status of the E2 Binding Site (E2BS) in the Upstream Regulatory Region (URR)[31]. The functional study has shown that E2BS1 can bind to transcription complex, resulting in enhanced E2 dependent promoter activity and increased expression of E6-E7 oncogene[32]. The viral URR is hypomethylated in the infected squamous cells which can replicate the viral genome and produce progeny viral particles[33]. However, the proximal Specificity Protein 1 (SP1) binding site of the E2BSs was in hypermethylated status in the superficial cell layers, it may be related to the down-regulation or even genes activation[32]. The methylation of HPV16 L1 affected E6/E7 messenger Ribonucleic Acid (mRNA) levels and Cervical Intraepithelial Neoplasia (CIN) 2+ was screened by detecting the methylation level of Human Papillomavirus 16 L1 (HPV16 L1) protein with sensitivity and specificity of 91.7 % and 59.5 %, respectively[34]. By detecting methylation of 12 HPV16 CpG sites, as a biomarker for the classification of HPV16-positive women, it was found that the methylation degree was correlated with the severity of cervical precancerous lesions and sensitivity to invasive cervical cancer[35]. Abnormal CpG methylation in the L1 and L2 regions of HPV18 and other high-risk HPV types (including HPV31/33/45/52/51/58) is associated with development of CIN [33,36]. In vitro studies have observed that upstream regulatory regioselectivity of type 16, 18 HPV methylation of CpG islands leads to a decrease in viral promoter transcriptional activity[37,38]. The HPV-16 proto-oncogene protein E7 was found to bind to the DNA Methyltransferase 1 (DNMT1) in vitro and in vivo, and increase the enzyme activity[39]. The interaction pattern of HPV infection with special site methylation can be used as a biomarker for early detection of the cervix and monitoring disease progression and prognosis.
The Diagnosed Value of ZNF582 Methylation in Cervical Cancer
ZNF582 and cervical cancer screening:
Based on the previous studies, it has been confirmed that the genes encoding Secreted Protein Acidic and Rich in Cysteine (SPARC) and Tissue Factor Pathway Inhibitor TFPI2) are highly methylated in cervical cancer cell lines. Huang et al. found that ZNF582 was highly methylated in cervical tumors through dual verification of cervical tumor cell smears and cervical cancer cell lines by array hybridization scanning and gene chip probes. The methylation frequency of ZNF582 is 100 % in cervical squamous cell carcinoma[8]. A study found that ZNF582 methylation was elevated in cervical exfoliated cells of patients with Cervical Adenocarcinoma (CAC) compared with the normal cervix (Odds Ratio (OR): 25.4, 95 % Confidence Interval (CI): 6.9-77.5), while the DNA methylation status of LIM Homeobox Transcription Factor 1 Alpha (LMX1A), NK6 Homeobox 1 (NKX6-1), Protein Tyrosine Phosphatase Receptor-Type R (PTPRR), Paired Box-1 (PAX1) and SRY-Box Transcription Factor 1 (SOX1) did not change[40]. Based on the above research, more and more studies have found that the methylation test of ZNF582 may be a biochemical indicator for screening of cervical cancer, improving screening rate, assisting diagnosis and evaluating treatment effects.
ZNF582 methylation associated with classification of cytological diagnosis of Low-Grade Squamous Intraepithelial Lesions (LSIL):
The traditional histopathological evaluation and high- risk HPV testing cannot determine the biological potential of LSIL[41,42]. Chang et al.[40] used ZNF582 as a shunt for LSIL, diagnosis and treatment biological markers. One multicenter prospective study detected ZNF582 gene methylation in cervical smears of 230 women with cytology diagnosed in 12 centers. The pathological biopsy results confirmed that 15 (6.5 %) of them were CIN3+. The sensitivity and specificity of HPV in CIN3+ were 80 % and 28 %, respectively (OR=1.6, 95 % CI: 0.4-5.8). However, when the ZNF methylation of methamphetamine (meth)-use index cutoff value predicted CIN3+, the sensitivity and specificity were 73 % and 71 %, respectively (OR: 6.8, 95 % CI: 2.1-22.1). It has been proved that ZNF methylation analysis may be an effective choice in the LSIL cytological diagnostic classification and its prediction result is superior to HPV detection[7].
Therefore, ZNF582 methylation may be a better molecular marker for the classification of cytological diagnosis of LSIL. ZNF582 methylation combined with HPV-16/18 was performed in 242 patients with vaginal Atypical Squamous Cells of Undetermined Significance (ASC-US) and atypical squamous cells, which cannot exclude HSIL (ASC-H). The results showed the sensitivity and specificity of ZNF582 single gene detection in predicting tumorigenesis, were 82.43 % and 76.79 %, respectively, and any positive ZNF582 or HPV16/18 could increase the sensitivity to 89.19 %. Combined screening of ZNF582 and HPV16/18 can increase cancer detection rates[10]. Therefore, they recommend ZNF582 methylation level detection and HPV-16/18 type detection for patients with cytological results of ASC-US or ASC-H. If any of ZNF582 methylation and HPV-16/18 type is positive, colposcopy is recommended. Vaginal microscopy is an invasive test and it is easy to aggravate the patient’s panic, while negative cytology can be performed every 3-6 mo, and this shunting measure reduces unnecessary colposcopy and cervical biopsy[43]. Shen-Gunther et al. found that HPV genotypes and DNA methylation of Adenylate Cyclase 8 (ADCY8), Cadherin 8 (CDH8), and ZNF582 are associated with cytological grades. Different grades of cervical cytology have unique molecular characteristics and can be converted into a multi-targeted “Pap smear” for clinical detection[44].
ZNF582 methylation enhanced the diagnostic efficacy of high-grade cervical cancer (CIN3):
Previously, the high-risk type of shunting method for HPV was cytologically tested. Tian et al. found the ZNF582 methylation combined with PAX1 or SOX1 was 100 % sensitive to cervical cancer diagnosis, but the positive rate of decline in the CIN3 category fell to between 62.86 % and 74.29 %. The study showed that HPV-16/18 genotyping combined with PAX1/ZNF582 methylation test can provide screening convenience for areas where colposcopy is not popular[45]. It is beneficial to reduce the anxiety of women with high- risk HPV infections. Because the combined screening rate of cervical cancer is higher than cytology, it can also reduce the unnecessary referral of cervical cancer patients. Liou et al. defined the methylation status of ZNF852 by the coefficient value in the regression formula and calculated the regression score in combination with the clinical characteristics (vaginal bleeding, tumor size, contact bleeding and presence or absence of HPV infection). They found that the combination of clinical characteristics and ZNF582 methylation threshold value for the diagnosis of CIN3+ category will get sensitivities and specificities of 85.4 % and 80.1 %, respectively, and the sensitivity to the diagnosis of cervical cancer was 100 %[9]. Recently, Li et al. conducted a meta-analysis in the Chinese population, comprising 1749 patients from seven studies and the results have shown that the combined sensitivity of ZNF582 methylation to the diagnosis of cervical intraepithelial neoplasia type III/more severe (CIN3+) patients was estimated to be 0.71 with a 95 % CI of 0.67 to 0.75 and the corresponding specificity is 0.81 with a 95 % CI of 0.79 to 0.83, the Area Under the Curve (AUC) is 0.85[46]. Stratified analysis proved that combined HPV and ZNF582 methylation detection have higher diagnostic accuracy than single HPV detection. Therefore, ZNF582 methylation is expected to become a biomarker to assist in cervical cancer screening. The new strategy of combining detection of HPV and ZNF582 methylation in cervical scrapings can improve diagnostic accuracy compared to single HPV detection. Our previous study had showed hypermethylation of the ZNF582 gene in cervical cancer.
Lately, we conducted a retrospective clinical investigation to investigate the diagnostic and predictive value of ZNF582 gene methylation in chemoradiotherapy sensitivity and prognosis of CAC, which compared the methylation of 219 cervical specimens comprising 189 cancer tissues (167 adenocarcinomas and 22 adenosquamous carcinomas) and 30 non-cancerous tissues (inflammation). The level of ZNF582 gene methylation in CAC tissues was higher than that in non-cancer tissues and the prognosis of patients with hypomethylation of ZNF582 gene was poor. We also found that the levels of ZNF582 methylation in Concomitant Chemoradiotherapy (CCRT) patients were lower than those in non-CCRT.
Hypomethylation was related to the high expression of ZNF582 protein and the overexpression of ZNF582 protein can increase the resistance of Henrietta Lacks (HeLa) cells to radiotherapy and chemotherapy. Overall, the abnormal methylation of ZNF582 may be a potential biomarker for the detection and prognosis monitoring of CAC. The overexpression of ZNF582 increases the chemoradiotherapy resistance of CAC[47].
Studies of ZNF582 Methylation in Other Cancers
In recent years, a lot of researchers have become increasingly enthusiastic about researches on methylation of ZNF582. Resources not only in cervical cancer but also in other cancers have been well published. In the table below, we have listed all the studies of ZNF582 except cervical cancer so far (Table 1)[48-53]. The results support ZNF582 hypermethylation as a biomarker for cancer screening.
| Cancer types | Conclusion | Samples | References |
|---|---|---|---|
| Oral cancer | ZNF582 hypermethylation-Associated with aggressive progression and poor prognosis of OSCC Biomarker for the detection of oral dysplasia and oral cancer |
Oral scrapings Oral epithelial cell samples collected by mouth rinse |
[48,49] |
| Esophageal Squamous Cell Carcinoma (ESCC) | ZNF582 hypermethylation-Biomarker for ESCC screening and diagnosis | Tumor tissues | [50,51] |
| Colorectal cancer | ZNF582 hypermethylation-Biomarker for the detection of colorectal cancer | Tumor tissues Bowel Lavage Fluid (BLF) |
[52] |
| Anal cancer | ZNF582 hypermethylation-Associated with anal carcinogenesis | Tumor tissues | [53] |
Table 1: ZNF582 Methylation is a Promising Biomarker for Cancer Detection
Discussion
Cervical cancer is a malignant tumor with high morbidity and mortality in women, and its 5 y survival rate is very low[54]. Epidemiological and molecular studies support the conclusion that persistent infection of high-risk HPV is a necessary condition for cervical cancer, but not the only prerequisite[55]. This promotes the development of HPV DNA testing and improves the screening strategy for cervical cancer in recent years.
In 2003, the United States Food and Drug Administration (USFDA) approved the combined use of Thrombin Clotting Time (TCT) and HPV- DNA Hybrid Capture 2 (HC2) testing for preliminary screening of women over the age of 30 y[56]. In 2005, the International Agency for Research on Cancer (IARC) of the World Health Organization recommended the use of high-risk HPV testing for preliminary screening of cervical cancer[57]. HPV infection may be temporary, even if the test is positive, the infection will be cleared over some time and only a few infected people will develop cancer after a long incubation period. Since there are over 20 HPV subtypes, it is not enough to detect a single HPV subtype. There is also an urgent need for additional genetic and epigenetic markers for the progression from precancerous lesions to invasive cancer.
At present, the detection of DNA methylation can be used as a biomarker for the detection of advanced CIN2/3 lesions[58,59]. Many methylation genes have been described as markers for HPV positive women, such as SOX1, PAX1, Septin 9 (SEPT9), DAPK1, Junctional Adhesion Molecule 3 (JAM3), Cell Adhesion Molecule 1 (CADM1) and Myelin and Lymphocyte protein (MAL)[60-64]. Early screening for malignant tumors was monitored by monitoring changes in ZNF582 methylation levels and finding appropriate cut-off values. In cervical cancer, ZNF582 may be used as one of the cytological diagnosis methods for the diagnosis and treatment of LSILs. Compared with colposcopy, cervical cytology methylation detection is more convenient and non-invasive. ZNF582 methylation level was detected in patients after radical surgery for Oral Squamous Cell Carcinoma (OSCC). It was found that ZNF582 methylation level increased 3-4 mo before cancer recurrence and elevated ZNF582 methylation level was positively correlated with poor prognosis. ZNF582 has the potential to be a sensitive marker for detecting tumor recurrence and disease progression[35]. Our current data shows that in the exfoliated cells of cervical cancer patients undergoing radical radiotherapy, methylation of ZNF582 decreases with the remission of the disease. We have found that ZNF582 can be used as a biomarker for predicting radiotherapy sensitivity. In the future, we will further study whether ZNF582 can be used as a prognostic indicator for chemoradiotherapy.
Author’s contributions:
Na Yiyuan Wu, Zhao Yi Liu, Chao Fang contributed for project development, data collection and manuscript writing. Na Yiyuan Wu and Chao Fang contributed regarding the idea of the article and manuscript corrections.
Funding:
This work was supported by the National Natural Science Foundation of China (82003867), Scientific Research project of Hunan Health Commission (202213015439, 20201487).
Conflict of interests:
The authors declared no conflict of interest.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google scholar] [PubMed]
- Wentzensen N, Walker J, Schiffman M, Yang HP, Zuna RE, Dunn ST, et al. Heterogeneity of high?grade cervical intraepithelial neoplasia related to HPV16: Implications for natural history and management. Int J Cancer 2013;132(1):148-54.
[Crossref] [Google scholar] [PubMed]
- Nessa A, Rashid MH, E?Ferdous N, Chowdhury A. Screening for and management of high?grade cervical intraepithelial neoplasia in Bangladesh: A cross?sectional study comparing two protocols. J Obstet Gynaecol Res 2013;39(2):564-71.
[Crossref] [Google scholar] [PubMed]
- Yang S, Wu Y, Wang S, Xu P, Deng Y, Wang M, et al. HPV?related methylation?based reclassification and risk stratification of cervical cancer. Mol Oncol 2020;14(9):2124-41.
[Crossref] [Google scholar] [PubMed]
- Dong L, Zhang L, Hu SY, Feng RM, Zhao XL, Zhang Q, et al. Risk stratification of HPV 16 DNA methylation combined with E6 oncoprotein in cervical cancer screening: A 10-year prospective cohort study. Clin Epigenetics 2020;12(1):1-11.
- Ma X, Liu J, Wang H, Jiang Y, Wan Y, Xia Y, et al. Identification of crucial aberrantly methylated and differentially expressed genes related to cervical cancer using an integrated bioinformatics analysis. Biosci Rep 2020;40(5).
[Crossref] [Google scholar] [PubMed]
- Lin H, Chen TC, Chang TC, Cheng YM, Chen CH, Chu TY, et al. Methylated ZNF582 gene as a marker for triage of women with Pap smear reporting low-grade squamous intraepithelial lesions-a Taiwanese Gynecologic Oncology Group (TGOG) study. Gynecol Oncol 2014;135(1):64-8.
[Crossref] [Google scholar] [PubMed]
- Huang RL, Chang CC, Su PH, Chen YC, Liao YP, Wang HC, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One 2012;7(7):e41060.
[Crossref] [Google scholar] [PubMed]
- Liou YL, Zhang TL, Yan T, Yeh CT, Kang YN, Cao L, et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics 2016;8(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Liou YL, Zhang Y, Liu Y, Cao L, Qin CZ, Zhang TL, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics 2015;7(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Waddington CH. The strategy of the genes: A discussion of some aspects of theoretical biology. 1st ed. London: London George Allen and Unwin; 1957.
- Szalmás A, Kónya J. Epigenetic alterations in cervical carcinogenesis. Semin Cancer Biol 2009;19(3):144-52.
[Crossref] [Google scholar] [PubMed]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature 1998;395(6697):89-93.
[Crossref] [Google scholar] [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358(11):1148-59.
[Crossref] [Google scholar] [PubMed]
- Greger V, Passarge E, Höpping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet 1989;83(2):155-8.
[Crossref] [Google scholar] [PubMed]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011;480(7378):490-5.
[Crossref] [Google scholar] [PubMed]
- Zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer 2002;2(5):342-50.
[Crossref] [Google scholar] [PubMed]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol 2001;11(1):39-46.
[Crossref] [Google scholar] [PubMed]
- Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 1989;245(4918):635-7.
[Crossref] [Google scholar] [PubMed]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform 2011;12(5):449-62.
[Crossref] [Google scholar] [PubMed]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010;17(1):13-27.
[Crossref] [Google scholar] [PubMed]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 2006;16(5):669-77.
[Crossref] [Google scholar] [PubMed]
- Saavedra KP, Brebi PM, Roa JC. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin Epigenetics 2012;4(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Snellenberg S, Cillessen SA, van Criekinge W, Bosch L, Meijer CJ, Snijders PJ, et al. Methylation-mediated repression of PRDM14 contributes to apoptosis evasion in HPV-positive cancers. Carcinogenesis 2014;35(11):2611-8.
[Crossref] [Google scholar] [PubMed]
- Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: Its relationship to clinical outcome. Mol Cancer 2003;2(1):1-2.
[Crossref] [Google scholar] [PubMed]
- Widschwendter A, Müller HM, Fiegl H, Ivarsson L, Wiedemair A, Müller-Holzner E, et al. DNA methylation in serum and tumors of cervical cancer patients. Clin Cancer Res 2004;10(2):565-71.
[Crossref] [Google scholar] [PubMed]
- Mani E, Medina LA, Isaac-Olive K, Duenas-Gonzalez A. Radiosensitization of cervical cancer cells with epigenetic drugs hydralazine and valproate. Eur J Gynaec Oncol 2014;35(2):2014.
[Google scholar] [PubMed]
- Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet 2007;370(9590):890-907.
[Crossref] [Google scholar] [PubMed]
- Nam KH, Kim YT, Kim SR, Kim SW, Kim JW, Lee MK, et al. Association between bacterial vaginosis and cervical intraepithelial neoplasia. J Gynecol Oncol 2009;20(1):39-43.
[Crossref] [Google scholar] [PubMed]
- Steenbergen RD, de Wilde J, Wilting SM, Brink AA, Snijders PJ, Meijer CJ. HPV-mediated transformation of the anogenital tract. J Clin Virol 2005;32(1):25-33.
[Crossref] [Google scholar] [PubMed]
- Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J Virol 2003;77(23):12450-9.
[Crossref] [Google scholar] [PubMed]
- Vinokurova S, von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS One 2011;6(9):e24451.
[Crossref] [Google scholar] [PubMed]
- Simanaviciene V, Popendikyte V, Gudleviciene Z, Zvirbliene A. Different DNA methylation pattern of HPV16, HPV18 and HPV51 genomes in asymptomatic HPV infection as compared to cervical neoplasia. Virology 2015;484:227-33.
[Crossref] [Google Scholar] [PubMed]
- Bergeron C, Ronco G, Reuschenbach M, Wentzensen N, Arbyn M, Stoler M, et al. The clinical impact of using p16INK4a immunochemistry in cervical histopathology and cytology: An update of recent developments. Int J Cancer 2015;136(12):2741-51.
[Crossref] [Google Scholar] [PubMed]
- Brandsma JL, Harigopal M, Kiviat NB, Sun Y, Deng Y, Zelterman D, et al. Methylation of twelve CpGs in human papillomavirus type 16 (HPV16) as an informative biomarker for the triage of women positive for HPV16 infection. Cancer Prev Res 2014;7(5):526-33.
[Crossref] [Google Scholar] [PubMed]
- Kalantari M, Osann K, Calleja-Macias IE, Kim S, Yan B, Jordan S, et al. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: Considerations for use as biomarkers of the progression of cervical neoplasia. Virology 2014;448:314-21.
[Crossref] [Google Scholar] [PubMed]
- Rösl F, Arab A, Klevenz B, Zur Hausen H. The effect of DNA methylation on gene regulation of human papillomaviruses. J Gen Virol 1993;74(5):791-801.
[Crossref] [Google Scholar] [PubMed]
- Stu?nkel W, Bernard HU. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol 1999;73(3):1918-30.
[Crossref] [Google Scholar] [PubMed]
- Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene 2007;26(11):1650-5.
[Crossref] [Google Scholar] [PubMed]
- Chang CC, Huang RL, Wang HC, Liao YP, Yu MH, Lai HC. High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1 and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol Cancer 2014;24(2):201-9.
[Crossref] [Google Scholar] [PubMed]
- Stoler MH. HPV testing is not useful for LSIL Triage-but stay tuned. Adv Anat Pathol 2001;8:160-4.
[Crossref] [Google Scholar] [PubMed]
- Reuschenbach M, Wentzensen N, Dijkstra MG, von Knebel Doeberitz M, Arbyn M. p16INK4a immunohistochemistry in cervical biopsy specimens: A systematic review and meta-analysis of the interobserver agreement. Am J Clin Pathol 2014;142(6):767-72.
[Crossref] [Google Scholar] [PubMed]
- Liou YL, Zhang Y, Liu Y, Cao L, Qin CZ, Zhang TL, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics 2015;7(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Shen-Gunther J, Wang CM, Poage GM, Lin CL, Perez L, Banks NA, et al. Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8 and ZNF582 as an integrated biomarker for high-grade cervical cytology. Clin Epigenetics 2016;8(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Tian Y, Wu NY, Liou YL, Yeh CT, Cao L, Kang YN, et al. Utility of gene methylation analysis, cytological examination, and HPV-16/18 genotyping in triage of high-risk human papilloma virus-positive women. Oncotarget 2017;8(37):62274-85.
[Crossref] [Google Scholar] [PubMed]
- Li N, He Y, Mi P, Hu Y. ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: A meta-analysis of related studies in Chinese population. Medicine 2019;98(6):e14297.
[Crossref] [Google Scholar] [PubMed]
- Wu NY, Zhang X, Chu T, Zhu S, Deng Y, Zhou Y, et al. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med 2019;7(14):328.
[Crossref] [Google Scholar] [PubMed]
- Cheng SJ, Chang CF, Ko HH, Liu YC, Peng HH, Wang HJ, et al. Hypermethylated ZNF582 and PAX1 genes in oral scrapings collected from cancer-adjacent normal oral mucosal sites are associated with aggressive progression and poor prognosis of oral cancer. Oral Oncol 2017;75:169-77.
[Crossref] [Google Scholar] [PubMed]
- Cheng SJ, Chang CF, Ko HH, Lee JJ, Chen HM, Wang HJ, et al. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck 2018;40(2):355-68.
[Crossref] [Google Scholar] [PubMed]
- Tang L, Liou YL, Wan ZR, Tang J, Zhou Y, Zhuang W, et al. Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomed Pharmacother 2019;120:109488.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Wang G, Tang J, Zhuang W, Wang LP, Liou YL, et al. DNA methylation status of PAX1 and ZNF582 in esophageal squamous cell carcinoma. Int J Environ Res Public Health 2017;14(2):216.
[Crossref] [Google Scholar] [PubMed]
- Harada T, Yamamoto E, Yamano HO, Nojima M, Maruyama R, Kumegawa K, et al. Analysis of DNA methylation in bowel lavage fluid for detection of colorectal cancer. Cancer Prev Res 2014;7(10):1002-10.
[Crossref] [Google Scholar] [PubMed]
- van der Zee RP, Richel O, van Noesel CJ, Novianti PW, Ciocanea-Teodorescu I, van Splunter AP, et al. Host cell deoxyribonucleic acid methylation markers for the detection of high-grade anal intraepithelial neoplasia and anal cancer. Clin Infect Dis 2019;68(7):1110-7.
[Crossref] [Google Scholar] [PubMed]
- Ginsburg O, Bray F, Coleman MP, Vanderpuye V, Eniu A, Kotha SR, et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017;389(10071):847-60.
[Crossref] [Google Scholar] [PubMed]
- Agorastos T, Chatzistamatiou K, Katsamagkas T, Koliopoulos G, Daponte A, Constantinidis T, et al. Primary screening for cervical cancer based on high-risk human papillomavirus (HPV) detection and HPV 16 and HPV 18 genotyping, in comparison to cytology. PLoS One 2015;10(3):e0119755.
[Crossref] [Google Scholar] [PubMed]
- Pan Z, Song Y, Zhe X, Wang W, Zhu J, Zheng W, et al. Screening for HPV infection in exfoliated cervical cells of women from different ethnic groups in Yili, Xinjiang, China. Sci Rep 2019;9(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Koliopoulos G, Nyaga VN, Santesso N, Bryant A, Martin?Hirsch PP, Mustafa RA, et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst Rev 2017(8):CD008587.
[Crossref] [Google Scholar] [PubMed]
- Boers A, Wang R, van Leeuwen RW, Klip HG, de Bock GH, Hollema H, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigenetics 2016;8(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KY, Ross JP. DNA methylation cancer biomarkers: Translation to the clinic. Front Genet 2019;10:1150.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Gao H, Tan HZ. SOX1 promoter hypermethylation as a potential biomarker for high-grade squamous intraepithelial neoplasia lesion and cervical carcinoma: A meta-analysis with trial sequential analysis. Front Genet 2020;11:633.
[Crossref] [Google Scholar] [PubMed]
- Zhao Z, Zhang X, Zhao X, Cai J, Wu NY, Wang J. SOX1 and PAX1 are hypermethylated in cervical adenocarcinoma and associated with better prognosis. Biomed Res Int 2020;2020:1-8.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Cui Z, Xiao Z, Hu M, Jiang C, Lin Y, et al. PAX1 and SOX1 methylation as an initial screening method for cervical cancer: A meta-analysis of individual studies in Asians. Ann Transl Med 2016;4(19):365.
[Crossref] [Google Scholar] [PubMed]
- Agodi A, Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M. DAPK1 promoter methylation and cervical cancer risk: A systematic review and a meta-analysis. PLoS One 2015;10(8):e0135078.
[Crossref] [Google Scholar] [PubMed]
- de Strooper LM, van Zummeren M, Steenbergen RD, Bleeker MC, Hesselink AT, Wisman GB, et al. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol 2014;67(12):1067-71.
[Crossref] [Google Scholar] [PubMed]
 Low ZNF582 TPM and
Low ZNF582 TPM and High ZNF582 TPM
High ZNF582 TPM



